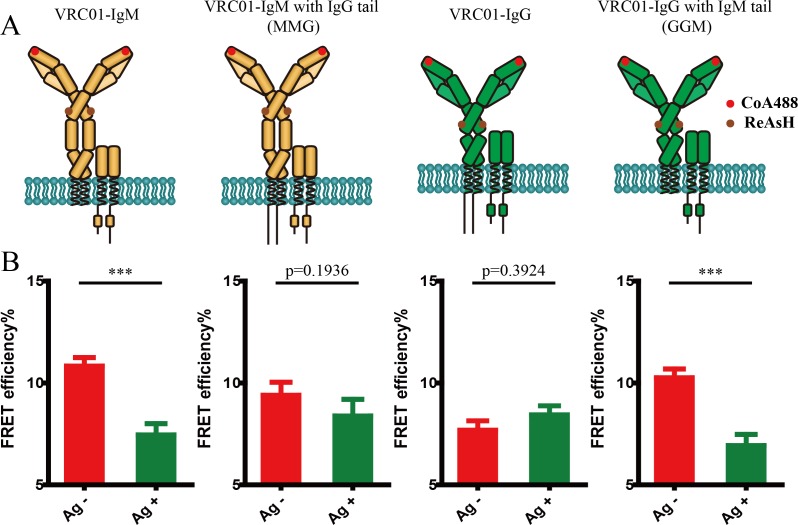
Figure 3. Conformational change within mIg heavy chain is dependent on the cytoplasmic tail.
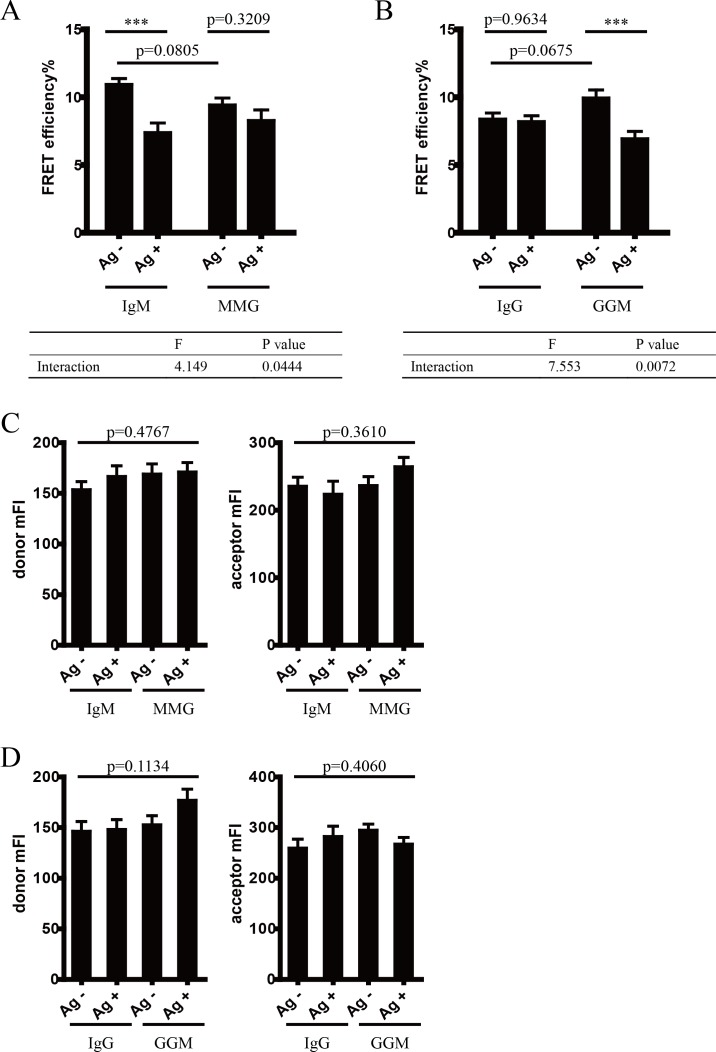
(A) Schematic illustration of different types of VRC01-BCR equipped with different cytoplasmic tail. FRET efficiency between CoA 488 label at N-terminus (red circle) and ReAsH at Cμ2 domain (or Cγ2 portion, brown circle) of dually tagged VRC01-BCR was examined. (B) Dequenching FRET to measure the FRET efficiency between CoA 488 and ReAsH in 293T cells expressing dually tagged VRC01-BCR activated by V51 monomer antigen. From left to right: IgM, MMG, IgG, GGM. Error bars represent mean ± SEM. Data are from at least 19 cells over two independent experiments. Two-tailed t-tests were used for the statistical comparisons. ***p<0.001.