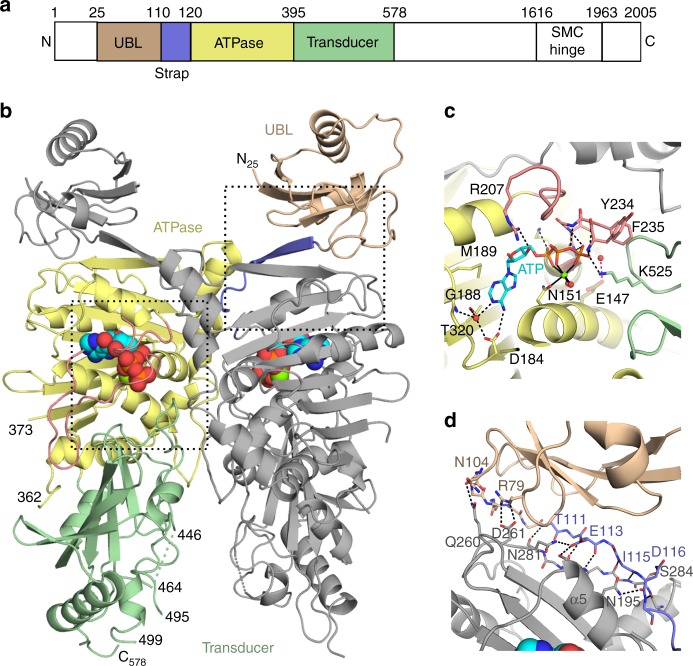
Fig. 1.
Dimeric structure of the N-terminal region of the SMCHD1 ATPase module. a Domain organization of full-length SMCHD1. b Cartoon of the crystal structure of the ATPase module (24–578aa). Residues in the pre-N ubiquitin-like domain (25–109aa), strap (110–120aa), GHKL-ATPase domain (121–395aa), and transducer domains (396–578aa) are colored wheat, lavender, pale-yellow and pale-green, respectively. The ATP molecule is colored cyan. c ATP binding to the active site. General location in the dimer is outlined with the bottom left box in panel b. ATP is shown in stick form alongside the catalytic Mg2+ (green) and coordinating water molecules (red spheres). Secondary structural elements are colored as in panels a and b. Hydrogen bonds to the ATP molecule and coordination interactions with the Mg2+ ion are shown as dashed and solid black lines, respectively. The catalytic glutamate (Glu147, transparent olive) has been modeled into the active site using the crystal structure of MORC2. The proposed hydrogen bond between Glu147 and the nucleophilic water is shown as a dotted red line. Both the ATP-lid (rose) and Lys525 from the switch-loop of the transducer domain form hydrogen bonds with the ATP phosphates. d Dimerization interactions of the UBL domain and strap. General location in the dimer is outlined with upper right box in panel b. There are extensive hydrogen bonds (black dotted lines) between the strap of one monomer (lavender) and the GHKL-ATPase domain (gray) of the other protomer. Additional interactions between the UBL domain (wheat) of one monomer and the GHKL-ATPase domain of the other monomer may also contribute to dimerization as constructs starting at residue 110 (excluding the UBL domain) do not dimerize