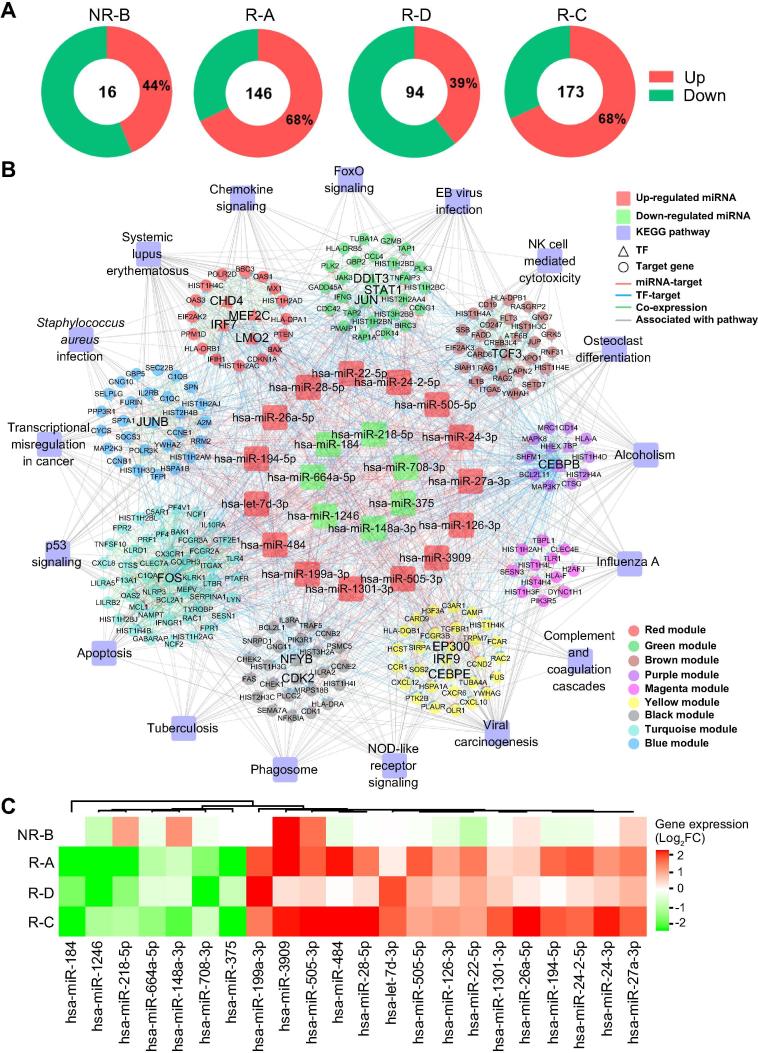
Figure 5.
The miRNA–TF–gene regulatory network involved in the CAR-T therapy
A. The distribution of DEMs across the four patients. The number of total DEGs and percentage of the up-regulated ones are provided. B. Important miRNA–TF–gene regulatory network. DEMs with a similar expression tendency in remissive patients were defined as important miRNAs, which together with TFs and their target genes formed the important miRNA-TF-gene regulatory network. The outer circle represents the KEGG pathways, the second circle represents the 9 co-expression modules depicted in different colors as shown in the legend box. The inner circle displays the 15 up-regulated miRNAs (red) and 7 down-regulated miRNAs (green) in the remissive samples. TFs and their target gene(s) are indicated in triangles and circles, respectively. Edges indicating miRNA–target regulation, TF–target regulation, co-expression between genes from different modules, and links between genes and pathways are shown in red, blue, green, and gray, respectively. C. Heatmap for DEMs with the same expression tendency in remissive patients. The color gradient from green to red indicates the fold change (log2 of post-VS-pre-CD19-CAR-T) of expression from low to high. DEM, differentially expressed miRNA.