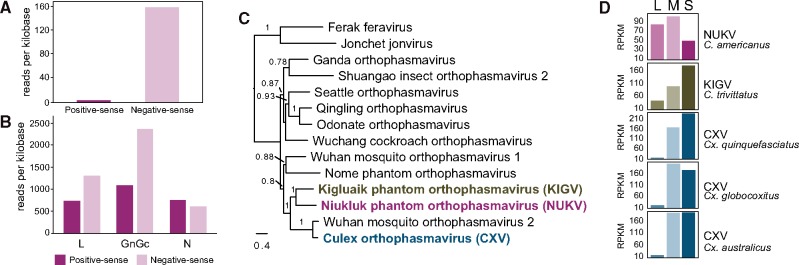
Figure 3.
Unusual genome segment abundance of NUKV in Chaoborus americanus. (A) Short-read mapping to the odin sequence shows that the gene is transcribed in antisense. (B) Short-reads mapping to the NUKV L and GnGc transcripts and genomes (negative sense) are more abundant than those of N—genome sequences appear to be disproportionately reduced. (C) A maximum likelihood phylogram of full-length L protein amino acid sequences shows evolutionary relationships between viruses in the family Phasmaviridae. KIGV and CXV are emphasized in bold typeface. Branches are labeled with SH-like approximate likelihood ratio test scores greater than 0.75. (D) Short-read mapping to full genome segment sequences reveals S segments are highly abundant in phasmaviruses related to NUKV (Ballinger et al. 2014; Shi et al. 2017).