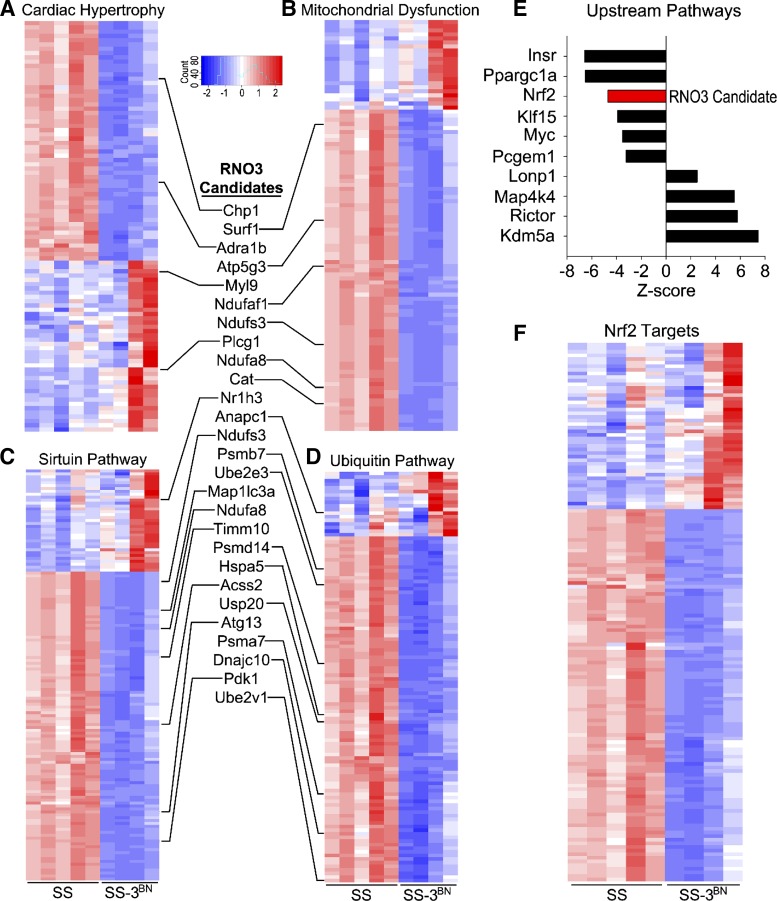
Fig. 8.
RNA-sequencing analysis of irradiated SS (Dahl salt-sensitive/Mcwi) and SS-3BN (substitution of rat chromosome 3 from the resistant Brown Norway rat strain onto the SS background) hearts. Total RNA was extracted, and RNA-sequencing was performed on RNA from the left ventricle of female SS and SS-3BN rats harvested 1 wk after 24 Gy of localized heart radiation or mock treatment (n = 4–5/group). Differential expression analysis was performed, and Ingenuity Pathway Analysis (IPA) gene network analysis identified the most significantly enriched gene ontologies to be cardiac hypertrophy (A), mitochondrial dysfunction (B), sirtuin signaling (C), and ubiquitin signaling (D), which included multiple candidate genes that reside on BN-derived rat chromosome 3 (RNO3; inset). E: analysis of the upstream molecular regulators of enriched pathways was also assessed by IPA, including the RNO3 candidate gene, Nrf2. F: differentially expressed Nrf2-target genes are significantly enriched following irradiation of SS and SS-3BN hearts.