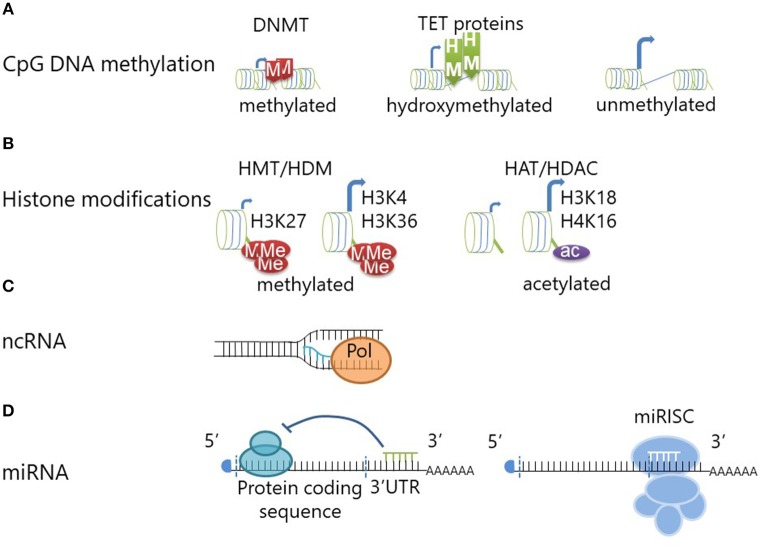
Figure 1.
Epigenetic modifications regulate gene transcription and translation. (A) DNA methyltransferase (DNMT) enzymes maintain or generate (de novo) DNA methylation at CpG dinucleotides. DNA methylation confers repression of gene expression through reduced transcription factor accessibility. DNA Hydroxymethylation is achieved through oxidation of methylated CpG DNA and mediated by Ten-eleven translocation methylcytosine dioxygenase (TET) proteins. DNA hydroxymethylation defines an “open” chromatin structure which allows for gene transcription (similar to unmethylated CpG DNA). (B) Histone methyltransferases (HMT) can add (one to three) methyl groups to histone amino termini. Depending on the exact molecular location and the degree of histone methylation, this can lead to chromatin compaction or decompaction. Methylation of histone H3 at lysine 27 (H3K27) will lead to chromatin compaction and transcriptional repression, while methylation at H3 lysine 4 (H3K4) and H3K36 mediates “opening” and increases transcription. Histone demethylases (HDM) can counteract this by removing the methyl groups. Histone acetylation is mediated by histone acetyltransferases (HAT) and can be reversed by histone deacetylases (HDAC). Histone acetylation is associated with chromatin decompaction and transcription of genes. (C) The transcription of non-coding RNA from intergenic or intronic regions can promote coding mRNA transcription by providing an open chromatin formation. (D) Short micro-RNAs (miRNA) can mediate transcriptional repression through inhibition of the ribosome when binding to the 3'UTR region of mRNAs. Furthermore, miRNAs can induce degradation of the mRNA through initiation of the miRISC complex.