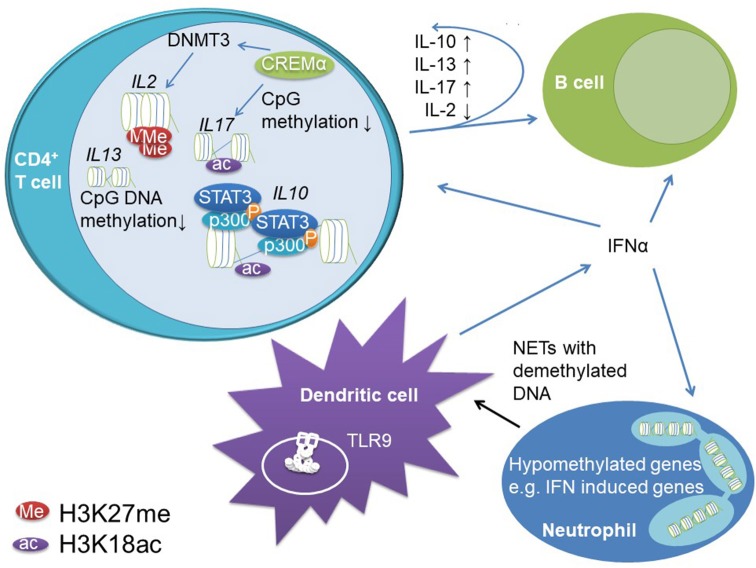
Figure 4.
Epigenetic mechanisms contribute to dysregulation of innate and adaptive immune responses in SLE. Reduced CpG DNA methylation at IL10 and IL13 regulatory regions allow for increased gene expression. STAT3 recruits to IL10 regulatory elements in the proximal promoter and an intrinsic enhancer. At these elements, STAT3 co-recruits the transcriptional co-activator p300 which has histone acetylase activity and supports chromatin decompaction through H3K18ac and increased gene expression. Increased IL-10 expression in T cells promotes B cell activity in SLE, while not affecting effector T cells (likely due to reduced IL-10 receptor expression on T cells from SLE patients). The transcription factor cAMP response element mediator (CREM)α promotes effector T cells in SLE. It induces H3K18 acetylation and CpG DNA demethylation across the IL17 gene cluster while recruiting DNMT3 to the IL2 locus instructing DNA methylation. Furthermore, CREMα co-recruits histone deacetylase (HDAC)1 to the IL2 gene, which results in decreased H3K18ac and stable gene silencing. Furthermore, B and T cells are stimulated by increased type I IFN (IFN-α and –β) expression in dendritic cells and neutrophils. Neutrophils exhibit reduced CpG DNA methylation of type I IFNs and associated genes. Dendritic cells are primed for type I IFN release through stimulation of endosomal TLR9 through augmented NETosis of neutrophils. Neutrophils from SLE patients release hypomethylated DNA, which binds to TLR9 more potently when compared to methylated DNA.