Figure 4.
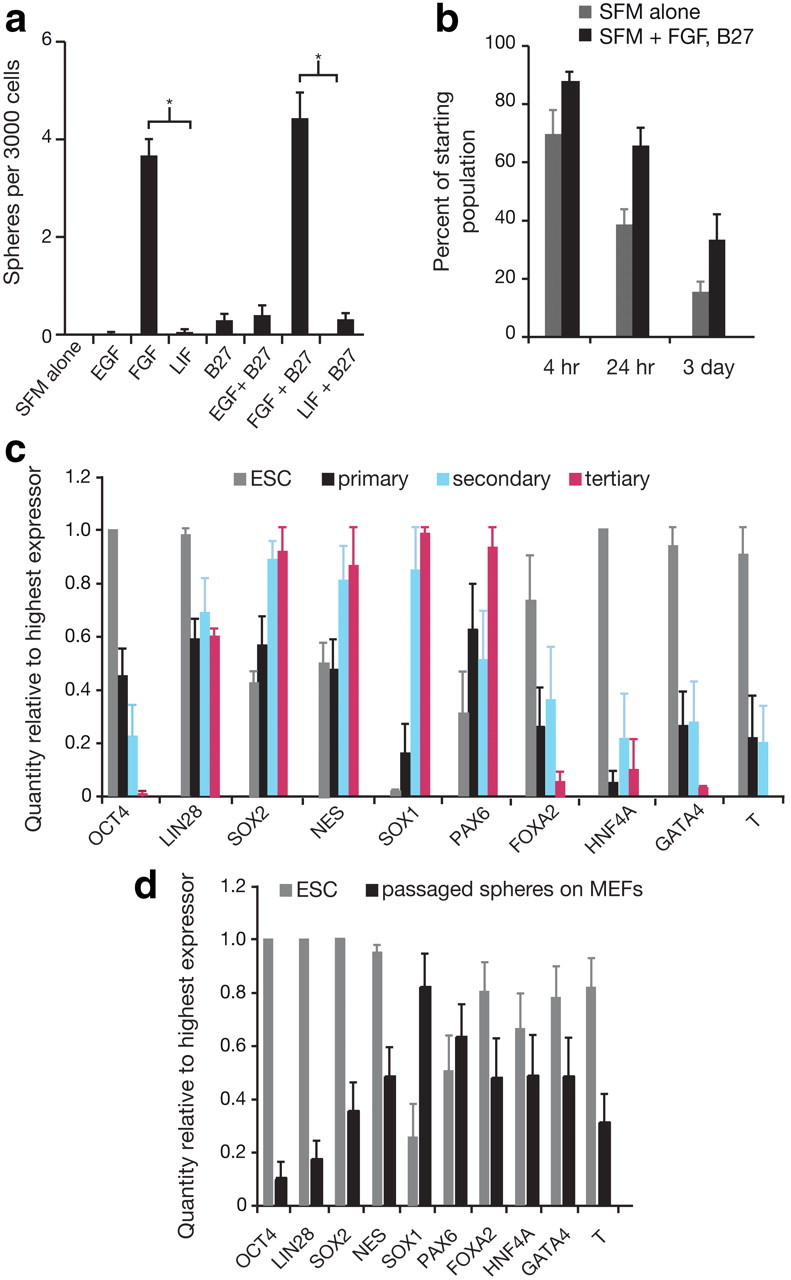
Primary neurosphere formation is dependent on FGF, and the addition of growth factors improves survival in SFM, but ROCK inhibitor influences cell fate. a, A comparison of culture conditions with addition of different growth factors reveals that the ability of single cells from hESC colonies to form neurospheres was reliant on the addition of FGF. One-way ANOVA showed a significant main effect of growth factors (F(7,31) = 56.46, p < 0.0001), while Bonferroni's multiple-comparison tests showed a significant difference between FGF and LIF (p < 0.05) and FGF+B27 compared with LIF+B27 (p < 0.05), illustrating that neurospheres are FGF dependent and not LIF dependent, n = 4. b, Single-cell survival in SFM versus SFM with growth factors represents the survivors as a percentage of starting cell populations. Two-way ANOVA showed that the survival of single cells in both conditions is diminished over time (F(2,30) = 42.40, p < 0.0001) and that the addition of growth factors to single cells in SFM improves survival (F(1,30) = 19.23, p = 0.0001), n = 3. c, Comparison of mRNA levels by Q-PCR showed that primary, secondary, and tertiary spheres cultured with ROCK inhibitor maintain expression of pluripotency (OCT4, LIN28) and non-neural genes (FOXA2, HNF4A, T and GATA4) unlike neurospheres from standard culture (Fig. 1k,l) that only express neural markers, n = 6. d, Q-PCR analysis of mRNA transcripts show that when ROCK inhibitor-cultured spheres are plated on MEFs in hESC maintenance conditions, ESC-like colonies form, and FOXA2 (p < 0.001), HNF4A (p < 0.001), and GATA4 (p < 0.001) expression is upregulated (by Bonferroni post-test comparison of primary, secondary, and tertiary spheres to passaged spheres on MEFs), n = 10. The data from c and d indicate that exposure to ROCK inhibitor prevents complete neuralization of hESCs in minimal conditions, resulting in colonies containing cells that are not stably committed to a neural lineage. Data are represented as mean ± SEM; for Q-PCR, the highest expressing sample (value of “1”) was used as the calibrator.
