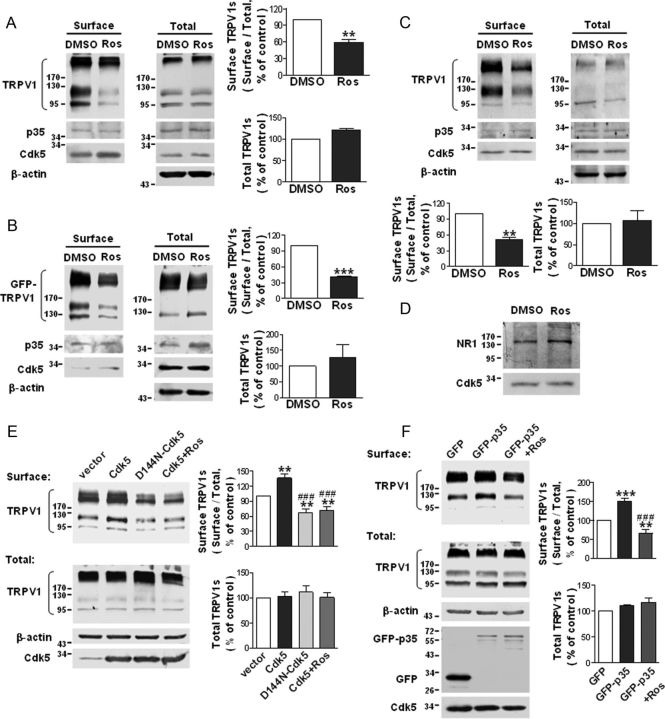
Figure 1.
Cdk5 plays a regulatory role in TRPV1 membrane localization. A–C, Roscovitine reduces the surface TRPV1 levels in TRPV1–CHO cells (A), F11 cells transfected with GFP–TRPV1 (B), and primary cultured DRG neurons (C). Cells were treated with 30 μm roscovitine for 5 h. The surface biotinylation assay was used to detect the amount of TRPV1 at the cell surface. The quantitative analyses show the ratio of surface to total TRPV1 normalized to the DMSO control and the total TRPV1 protein levels normalized to the DMSO control. Data were analyzed by two-tailed paired t test. **p < 0.01, ***p < 0.001 compared with DMSO control group, n = 3–4. Error bars indicate SEM. Ros, Roscovitine. D, The levels of surface NR1 subunits of the NMDA receptor in F11 cells are not reduced by roscovitine treatment (30 μm, 5 h). E, Overexpression of Cdk5 increases and overexpression of D144N–Cdk5 decreases the levels of surface TRPV1 in TRPV1–CHO cells. Cells were transfected with pcDNA3.1(+)–vector, pcDNA3.1(+)–Cdk5, or pcDNA3.1(+)–D144N-Cdk5. For the Cdk5+Ros group, cells expressing Cdk5 were treated with roscovitine (30 μm) for 5 h. Quantitative analysis show the ratio of surface to total TRPV1 normalized to the vector control (top) and the total TRPV1 protein levels normalized to the vector control (bottom). Data were analyzed by one-way ANOVA, followed by Newman–Keuls post hoc tests. **p < 0.01 compared with the vector group; ###p < 0.001 compared with the Cdk5 group, n = 5–6. Error bars indicate SEM. F, Overexpression of p35 increases the levels of surface TRPV1 in TRPV1–CHO cells. Cells were transfected with pEGFP–C2–p35 or pEGFP–C2–vector. For the GFP-p35+Ros group, cells expressing GFP–p35 were treated with 30 μm roscovitine for 5 h. Quantitative analysis of the ratio of surface to total TRPV1 normalized to the GFP control (top) and the total TRPV1 protein levels normalized to the GFP control (bottom) are shown. Data were analyzed by one-way ANOVA, followed by Newman–Keuls post hoc tests. **p < 0.01, ***p < 0.001 compared with the GFP group; ###p < 0.001 compared with the GFP–p35 group, n = 4–5. Error bars indicate SEM.