Figure 3.
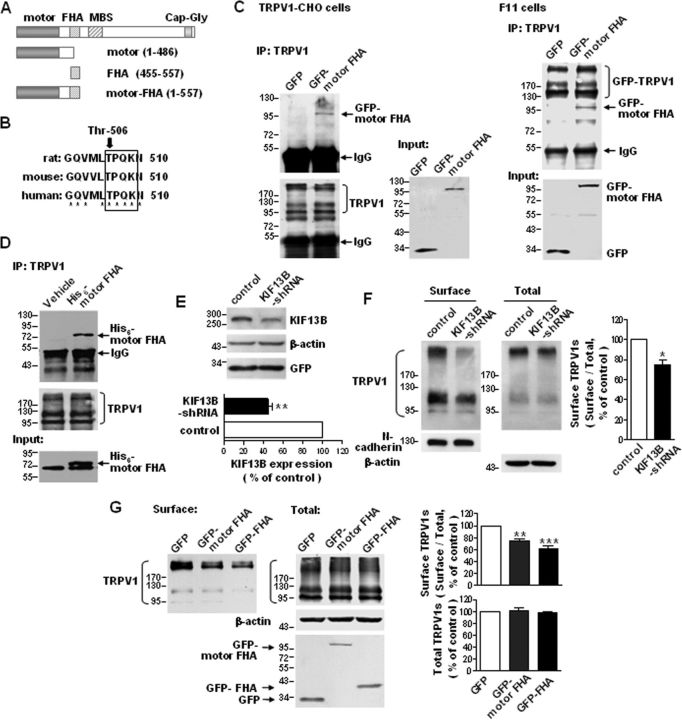
KIF13B transports TRPV1 to the cell surface. A, Schematic diagram of the domain structure of KIF13B/GAKIN and the constructs used in the present study. B, Comparison of the rat, mouse, and human KIF13B sequences containing the putative phosphorylation site of Cdk5. Threonine-506 is conserved across the species shown here. * indicates identical residues. C, Co-IP of TRPV1 and GFP–motor FHA. TRPV1–CHO cells (left) were transfected with pEGFP–C2–motor FHA or pEGFP–C2–vector, and F11 cells (right) were cotransfected with GFP–TRPV1 and pEGFP–C2–motor FHA or pEGFP–C2–vector. TRPV1s were immunoprecipitated with an anti-TRPV1 antibody, and the immunoblots were probed with an anti-GFP antibody. Lysates of cells expressing GFP were used as controls. Inputs represent the direct immunoblotting of the lysates. D, In vitro pull-down assay. TRPV1s were immunoprecipitated from cell lysates containing His6–motor FHA, which was expressed in bacteria. The immunoblots were detected with an anti-His antibody. E, Western blot analysis demonstrating an efficient decrease in KIF13B protein levels in primary cultured DRG neurons after infection with LVs expressing KIF13B shRNAs. **p < 0.01, n = 3, two-tailed paired t test. Error bar indicates SEM. F, Effect of KIF13B knockdown on the surface targeting of TRPV1s in primary cultured DRG neurons. The amount of membrane localized TRPV1 was detected with a surface biotinylation assay. Detection of N-cadherin was used as a control for the equal loading of biotinylated protein. The quantitative analysis shows the ratio of surface to total TRPV1 normalized to the control. *p < 0.05, n = 3, two-tailed paired t test. Error bar indicates SEM. G, Effect of overexpression of GFP–motor FHA or GFP–FHA on the surface targeting of TRPV1s in TRPV1–CHO cells. The top shows the quantitative analysis of the ratio of surface to total TRPV1 normalized to the GFP control. The bottom shows the total TRPV1 protein levels normalized to the GFP control. **p < 0.01, ***p < 0.001 compared with the GFP control, n = 3, one-way ANOVA, followed by Newman–Keuls post hoc tests. Error bars indicate SEM. Experiments in C and D were repeated at least three times.