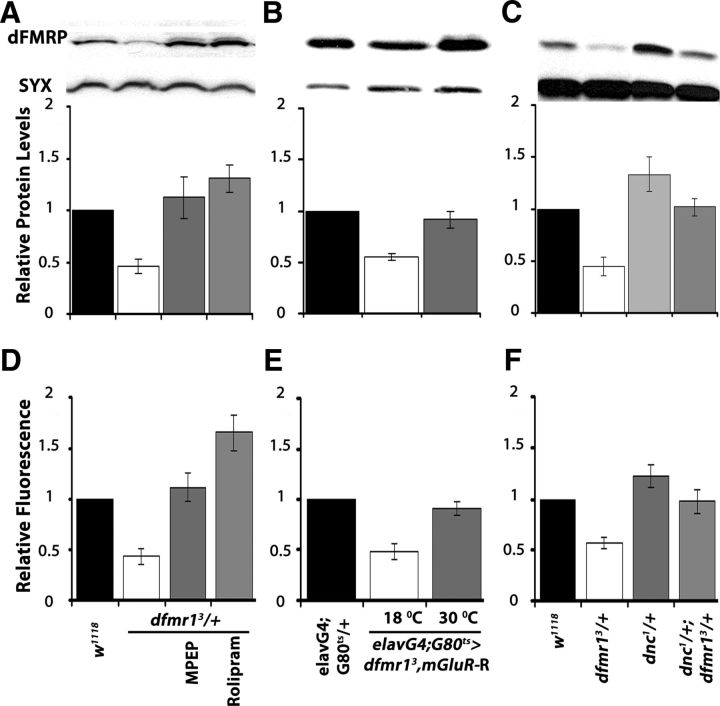
Figure 7.
Genetic and pharmacological manipulation altering dFMRP levels also affect the levels cAMP. A–C, Representative blots probing dFMRP levels in the indicated genotypes (top) and quantitative representation of the means ± SEM of the results relative to Syntaxin (SYX) which served as loading control in three such independent blots per experimental set. The genotypes of each group are shown on the bottom. Relative dFMRP levels in control strains (black bars) were arbitrarily fixed as 1 and were used to compare the levels in the other genotypes (Dunnett's tests). The significant (p < 0.0001) 50% decrease of the protein in dfmr13/+ is evident (open bars). A, In contrast, MPEP and Rolipram administration restored dFMRP to control levels (p = 0.12 and p = 0.002, respectively). B, Similarly, DmGluRA abrogation resulted in significantly higher (p < 0.0001) dFMRP levels. Animals were heterozygous for all transgenes and Gal4 and Gal80ts as indicated. C, dFMRP levels are significantly elevated in dnc1/+ over that in controls (p < 0.0001), but the levels in dnc1/+; dfmr13/+ animals are not (p = 0.67). D–F, cAMP levels measured in head lysates of the indicated genotypes using the Bridge-IR Fluorescent assay as detailed in Materials and Methods. cAMP levels in controls were arbitrarily designated as 1, and all other measurements are presented relative to that. The means ± SEM of three independent measurements (each the mean of a duplicate) are shown. D, Following a significant result from ANOVA (p < 0.0001), Dunnett's tests showed that cAMP levels in dfmr13/+ are significantly different (p < 0.0001) than controls, but they are restored to normal levels upon MPEP treatment (p = 0.25). As expected, following Rolipram treatment cAMP levels are significantly higher (p < 0.0001) than in controls. E, Conditional abrogation of DmGluRA in dfmr13/+ restores cAMP to control levels (p = 0.45), whereas the nucleotide levels are significantly lower (p < 0.0001) than controls if the transgene remains uninduced. F, cAMP levels are significantly higher (p < 0.0001) than controls in dnc1 heterozygotes, but in dnc1/+; dfmr13/+ flies are not significantly different from in controls (p = 0.06).