Figure 8.
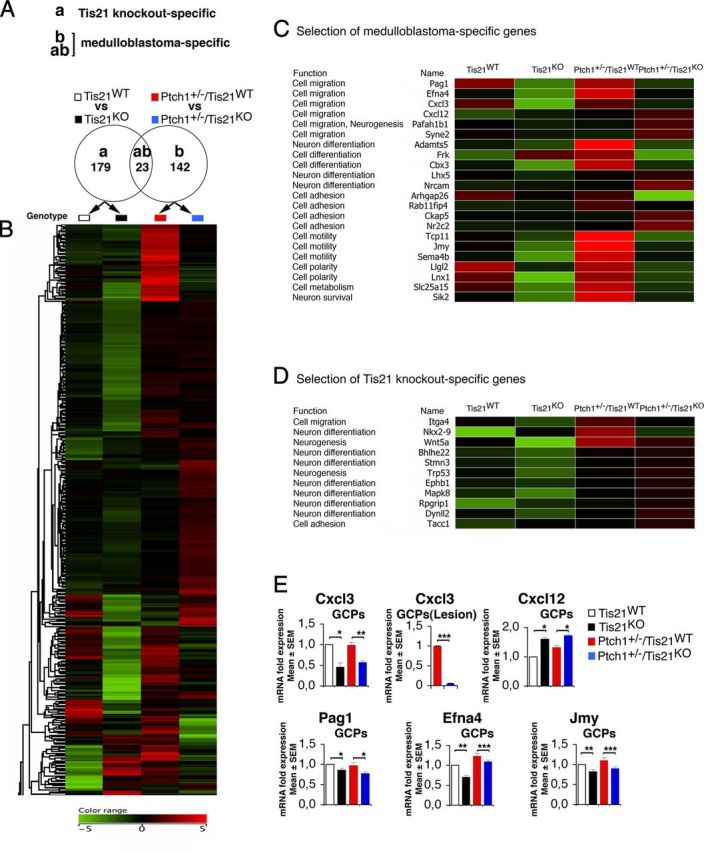
Medulloblastoma-specific gene expression in Patched1 heterozygous/Tis21-null model. A, As shown in the Venn diagram, microarray analysis of GCPs in P7 mice identified 179 Tis21 knock-out-specific genes, whose expression significantly differed only in pairwise comparison (a): Patched1 wild-type/Tis21-null versus wild type (Tis21KO vs Tis21WT). Likewise, 165 medulloblastoma-specific genes were identified whose expression significantly differed in pairwise comparison (b): Patched1 heterozygous/Tis21-null versus Patched1 heterozygous/Tis21 wild-type (Ptch1+/−/Tis21KO vs Ptch1+/−/Tis21WT). Twenty-three genes significantly differed in both comparisons (a) and (b). B, A hierarchical clustering algorithm (similarity measure, euclidean; linkage rule, centroid) was used to order the 344 genes differentially expressed in both pairwise comparisons (a) and (b). Colors indicate high expression (red) to low expression (green), corresponding to changes of normalized values relative to the median value. C, Twenty-two medulloblastoma-specific genes are involved in cell migration, differentiation, or adhesion processes. D, Eleven Tis21 knock-out-specific genes are involved in cell migration or differentiation processes. E, The differential expression of five neuronal medulloblastoma-specific genes in GCPs from the EGL at P7 or from lesions (GFP+) at 6 weeks was confirmed by real-time PCR, analyzing mRNA fold expression relative to wild-type mice (set to unity). Mean ± SEM fold increases are from three independent experiments. TBP and GAPDH were used to normalize data. *p < 0.05, **p < 0.01, or ***p < 0.001, Student's t test.
