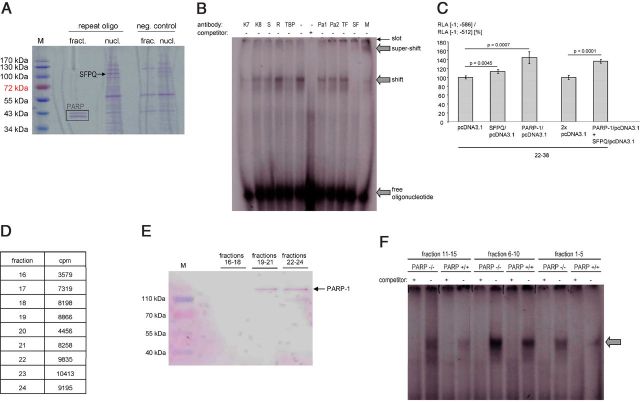
Figure 4.
Analyses of interactions between SFPQ and PARP-1 with the ECE-1c promoter. A, SDS-PAGE (Coomassie staining) after DNA affinity chromatography using the ECE-1c promoter-derived oligonucleotide 5′-tcttacacacacgcgcgcgcgcgcgcgcacacacacacacacacacacacacacacacacacaagcag-3′ (repeat oligo) and proteins from KELLY cells (nucl., total nuclear proteins; fract., gel-filtration fraction no. 28 of nuclear proteins). Beads without oligonucleotides served as negative (neg.) control. M, molecular weight marker; PARP, PARP-1 (degradation) fragments as identified by MS. B, EMSA using nuclear proteins from KELLY cells after gel filtration and the oligonucleotide as in A. For supershift analyses antibodies against Ku-70 (K7), Ku-86 (K8), Sam68 (S), RecQ1 (R), TBP, PARP-1 (Pa1, Pa2), TFII-I (TF), or SFPQ (SF) were used. These proteins represent candidates for ECE-1c promoter binding based on, for example, MS data. The specificity of the SFPQ supershift is further illustrated by the disappearance of the shifted band. M, mixture of anti-PARP-1, anti-TFII-I, and anti-SFPQ antibodies. C, The luciferase promoter constructs [−1; −512] (i.e., without the repeat) or [−1; −586] (comprising a human repeat consisting of 6 bp(CA)-22 bp(CpG)-38 bp(CA)) and two expression vectors encoding PARP-1 and SFPQ, respectively, were transiently (co)transfected into KELLY cells (n = 4 wells). Transient transfections of an insertless expression vector (pcDNA3.1) served as negative controls. The ratio (RLA [−1; −586] construct/RLA [−1; −512] construct) indicates the contribution of the microsatellite to the total promoter activity. D, Confirmation of the PARP-1-DNA interaction by gel filtration. A double-stranded, 32P-labeled deoxyoligonucleotide derived from the human ECE-1c promoter repeat (5′-tcttacacacacgcgcgcgcgcgcgcgcacacacacacacacacacacacacacacacacacaagcag-3′) was incubated with nuclear proteins of KELLY cells followed by gel filtration. Radioactivity was measured in all 40 gel filtration fractions and proteins were precipitated by trichloroacetic acid in fractions 16—24, which exhibited the highest counts per minute (cpm) values. E, Western blotting after gel filtration. Fractions from D were pooled as indicated followed by Western blotting using an anti-PARP-1 antibody. M, molecular weight marker. F, EMSA using PARP-1-deficient cells. Nuclear proteins were extracted from MEFs of homozygous PARP-1 knock-out mice (PARP−/−) or respective wild-type controls (PARP+/+) followed by gel filtration. Subsequent EMSA was performed with the radiolabeled oligonucleotide 5′-tcttacacacacgcgcgcgcgcgcgcgcacacacacacacacacacacacacacacacacacaagcag-3′ derived from the human ECE-1c promoter. The arrow indicates a specific, i.e., competeable, [CpG]m[CA]n-transcription factor complex.