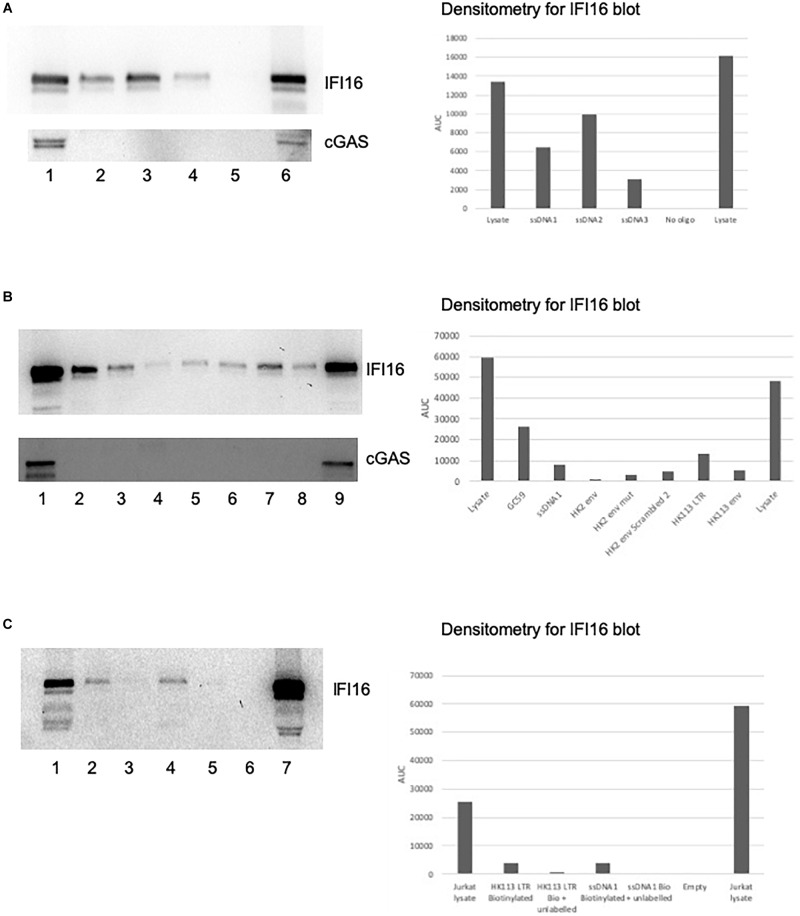
FIGURE 2.
IFI16 binds to oligos from HIV-1 and HERV-K. Pre-cleared Jurkat cell lysates were incubated with biotin-labeled oligos, as described in the section “Materials and Methods.” The pulldowns were analyzed by SDS-PAGE and western blotting first for IFI16, followed by reprobing the same blot for cGAS. (A) The ssDNA 1, 2 and 3 oligos (Jakobsen et al., 2013) were examined for their interaction with IFI16 (top panel) and cGAS (bottom panel). The lanes were loaded with input lysate or pulldowns with oligos as indicated: (1) input lysate, (2) ssDNA1, (3) ssDNA2, (4) ssDNA3, (5) no oligo control and (6) repeat of the input lysate. (B) A HK2 oligo from the consensus sequence, as well as the HK113 sequence, were analyzed for interaction with IFI16 (top panel) and cGAS (bottom panel). The lanes were loaded with input lysate or pulldowns with oligos as indicated: (1) input lysate, (2) GC59, (3) ssDNA1, (4) HK2 env, (5) HK2 env mut, (6) HK2 env scrambled 2, (7) HK113 LTR, (8) HK113 env and (9) a positive input lysate from a previous experiment as a blotting control. (C) In this competition assay, the biotinylated ssDNA1 and HK113 LTR oligos were used in pulldowns with and without an untagged version of the same oligo. The lanes were loaded as follows: (1) input lysate, (2) HK113 LTR biotinylated oligo only, (3) HK113 LTR biotinylated and unlabeled oligos, (4) ssDNA1 biotinylated oligo only, (5) ssDNA1 biotinylated and unlabeled oligos, (6) empty lane and (7) repeat of the input lysate. Densitometry was performed on the IFI16 blots by measuring the area under the curve (AUC) with Image J.