Fig. 3.
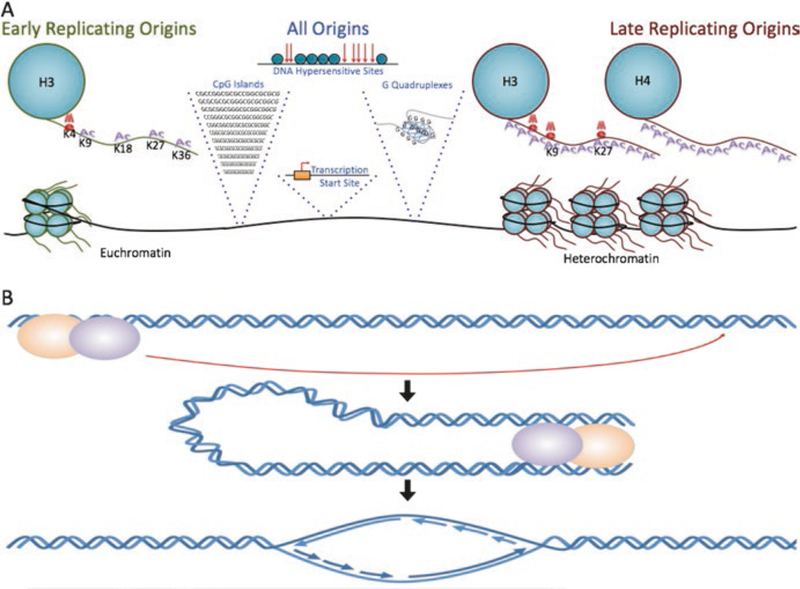
(a) Metazoan replication origins share several sequence features. Origins generally associate with regions that exhibit strand asymmetry, CpG islands, G-quadruplexes, transcription start sites, origin G-rich-repeated elements (OGREs), and regions of DNase hypersensitivity. In agreement, local histone modifications correlate with and can determine replication origin locations and timing. Early replicating regions associate with H3K4me1/2/3, H3K9ac, H3K18ac, H3K36me3, and H3K27ac. These histone modifications also associate with open chromatin and are enriched in moderately active transcription start sites. Late replicating regions tend to associate with H3 and H4 hypoacetylation, H3K9 and H3K27 methylation, and are found in heterochromatic regions. (b) Distal DNA sequences affect origin activity and transcriptional through long-distance interactions. Such interactions can be mediated via protein interaction with enhancers and locus control regions, by chromatin remodeling factors and transcriptional activators that bind enhancers and locus control regions, and by long noncoding RNAs
