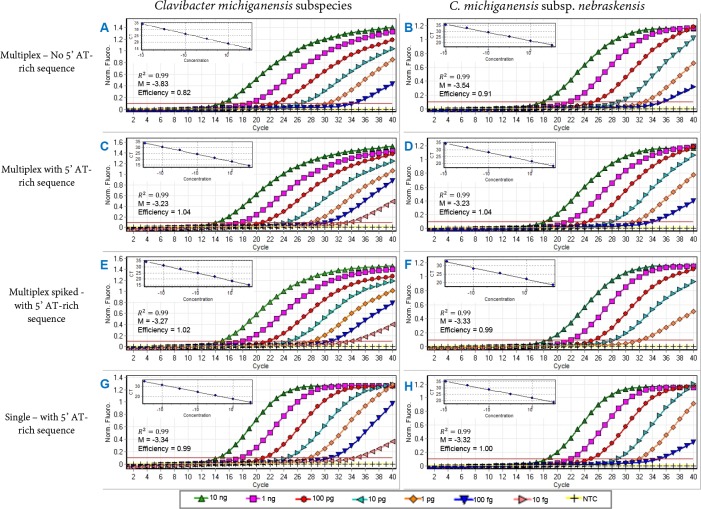
Fig 4. Effect of 5’ AT-rich flap on TaqMan real-time qPCR sensitivity assays performed using ten-fold serially diluted genomic DNA.
(A) and (B). TaqMan real-time qPCR assays with individual primer set harboring no complementary 5’ AT-rich sequences. (A) represents the detection C. michiganensis; and (B) represents the subspecific detection of C. m. subsp. nebraskensis; (C, D, E, F and G) are multiplex and single reactions after adding the non-complementary 5’ AT-rich sequences. (C) and (D) multiplex reactions for simultaneous detection of all subspecies of C. michiganensis and of C. m. subsp. nebraskensis, respectively. (E) and (F) multiplex reactions conducted for simultaneous detection of C. michiganensis subspecies and of C. m. subsp. nebraskensis, respectively, after adding 1 μl of healthy plant corn DNA to each 10-fold serially diluted sensitivity reactions in order to simulate natural infection. (G) and (H) TaqMan real-time qPCR assays performed with individual primer and probe sets to detect C. michiganensis subspecies and C. m. subsp. nebraskensis, respectively. All assays were conducted from 10 ng to 10 fg serially diluted genomic DNA of C. m. subsp. nebraskensis (strain A6206) using Rotorgene.