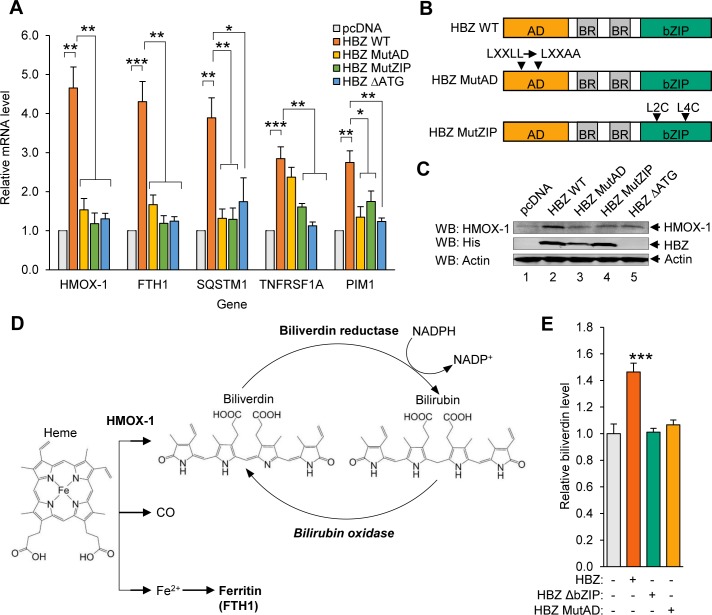
Fig 1. A subset of oxidative stress-induced genes that includes HMOX1 is upregulated in HBZ-expressing cells.
(A) A set of oxidative stress-response genes exhibit increased expression in the presence of HBZ. qRT-PCR was used to quantify relative mRNA levels for the indicated genes in HeLa clonal cell lines expressing wild-type HBZ (HBZ WT), an activation domain mutant of HBZ (HBZ MutAD), a leucine zipper domain mutant of HBZ (HBZ MutZIP), or a translational mutant of HBZ (HBZ ΔATG), and in a cell line containing the empty expression vector (pcDNA). HMOX1 values are averages from three independent experiments; FTH1, SQSTM1, TNFRSF1A, and PIM1 values are averages from four independent experiments. Data were normalized to the pcDNA sample (set to 1). Error bars represent SEM (two-tailed Student’s t-test, *p≤0.05, **p≤0.01, ***p≤0.001). (B) The schematics show the domains of HBZ and the mutants used in this study. Wild type HBZ (HBZ WT) consists of an N-terminal activation domain (AD), centrally located basic regions (BR), and a C-terminal basic leucine zipper domain (bZIP). HBZ MutAD contains LL→AA substitutions in two LXXLL motifs that render the AD defective. HBZ MutZIP contains L→C substitutions in the second and fourth heptad repeats of the ZIP domain, which disrupts dimerization with other bZIP factors. (C) HBZ upregulates HMOX-1 expression. Levels of the indicated proteins were evaluated in 30 μg of whole cell extract from each of the HeLa cell lines. The indicated antibodies were used for the Western blot (WB) analysis. (D) The schematic shows heme metabolism by HMOX-1. HMOX-1 cleaves the protoporphyrin ring of heme, creating biliverdin, carbon monoxide (CO), and free ferrous iron (Fe2+). Ferrous iron is scavenged by Ferritin (FTL/FTH1), and biliverdin reductase converts biliverdin to bilirubin. Bilirubin can be converted back to biliverdin by bilirubin oxidase. (E) Higher HMOX-1 levels in HBZ-expressing cells is associated with increased HMOX-1 enzymatic activity in these cells. Biliverdin production was quantified as a measure of HMOX enzyme activity in the indicated HeLa cell lines. Cells were homogenized and lysates were incubated with the HMOX substrate, hemin, and bilirubin oxidase as described in the Materials and Methods. Data are an average of three independent experiments. Error bars represent SEM (two-tailed Student’s t-test, **p≤0.01).