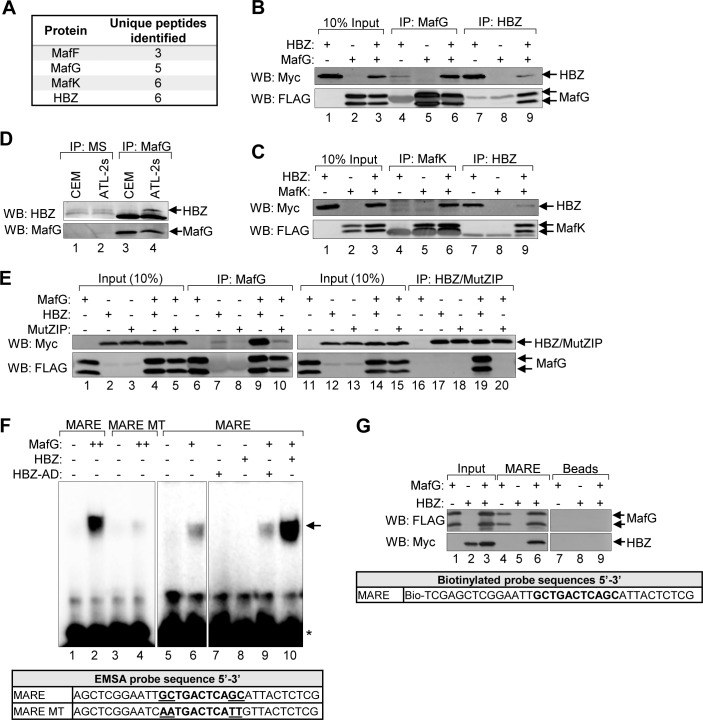
Fig 4. HBZ interacts with the small Mafs to form a DNA-bound complex at MAREs.
(A) A proteomic analysis identified all three small Mafs (MafF, MafG, and MafK) as potential HBZ-binding partners. A FLAG antibody was used to immunoprecipitate proteins from whole cell extracts prepared from a HeLa clonal cell line expressing HBZ with a C-terminal FLAG tag or a cell line containing the empty vector. Immunoprecipitates were analyzed by LC-MS/MS. Protein identifications were accepted at a false discovery rate of <1%, and a probability of ≥95%, and a maximum of one missed cleavage [68]. The number of unique peptides corresponding to each small Maf identified are shown. Data were obtained from a single experiment. (B and C) HBZ interacts with the small Mafs in transfected HEK 293T cells. Cells were transfected with 6 μg pcDNA-HBZ-Myc-His and/or 6 μg pCMV-MafG-FLAG or pCMV-MafK-FLAG (adjusted to 12 μg of total plasmid with the empty vector). HBZ and the small Mafs were immunoprecipitated using anti-Myc (IP: Myc) and anti-FLAG (IP: FLAG) antibodies, respectively. Immunoprecipitates and ten percent of the whole cell extract inputs were analyzed by Western blot using the antibodies indicated. (D) Endogenous MafG and HBZ interact in ATL cells. MafG (IP:MafG) was immunoprecipitated from CEM or ATL-2s whole cell extracts. Mouse serum (IP:MS) was used as a negative control for immunoprecipitation. Immunoprecipitates were analyzed by Western blot using the antibodies indicated. (E) The small Maf/HBZ interaction requires the ZIP domain of HBZ. HEK 293T cells were transfected with 1 μg pCMV-MafG-FLAG and/or 6 μg pcDNA-HBZ-Myc-His or 6 μg pcDNA-HBZ-MutZIP-Myc-His (adjusted to 12 μg of total plasmid with the empty vector). HBZ/MutZIP and the small Mafs were immunoprecipitated using anti-Myc (IP: Myc) and anti-FLAG (IP: FLAG) antibodies, respectively. Immunoprecipitates and ten percent of the whole cell extract inputs were analyzed by Western blot using the antibodies indicated. (F) HBZ augments formation of a MafG/MARE complex in vitro. In EMSAs, the indicated combination of recombinant, purified MafG-His (0.5 nM or 1 nM), GST-HBZ (50 nM), and GST-HBZ-AD (50 nM) were combined with a DNA probe containing the consensus T-MARE sequence or one containing AA mutations in the flanking GC boxes (MARE MT; sequences shown below) prior to electrophoretic separation of complexes. The panels shown are from the same gel/same scanned image (identical threshold adjustment). The arrow denotes the shifted protein/DNA complex; the asterisk denotes unbound probe. (G) In the presence of MafG, HBZ binds DNA containing the T-MARE sequence. Nuclear extracts prepared from HEK 293T cells transfected with pcDNA-HBZ-Myc-His and/or pCMV-MafG-FLAG were combined with a biotinylated DNA probe containing the T-MARE sequence coupled to streptavidin beads or with streptavidin beads alone (Beads). Bound proteins and 10% of the nuclear extract inputs were analyzed by Western blot using the antibodies indicated.