Figure 8.
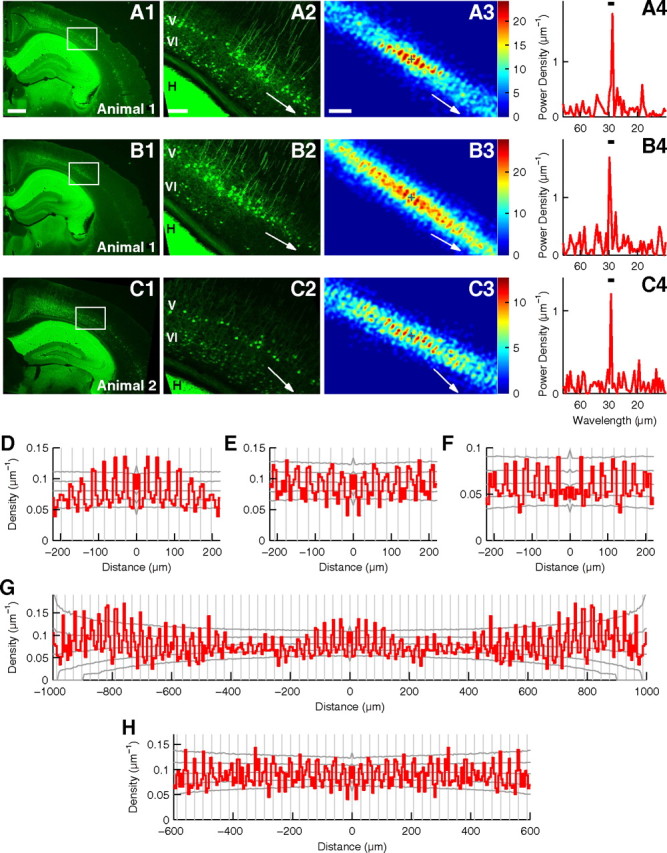
Periodic arrangement of EGFP(+) cells in the visual cortex of adult Crym-BAC-EGFP mice. A, Analysis of a coronal slice of the visual cortex of a P41 Crym-BAC-EGFP mouse stained for EGFP protein. A1, Low-magnification photograph. Scale bar, (in A1) A1, B1, and C1, 500 μm. A2, High-magnification photograph of the area indicated by a rectangle in A1. This area includes the binocular region, and it was used for periodicity analysis. White arrow denotes the orientation of periodicity. H, Hippocampus; V and VI, layers V and VI, respectively, for A2, B2, and C2. Scale bar, (in A2) A2, B2, and C2, 100 μm. A3, Histogram of relative positions of EGFP(+) cells in layer V. The origin is shown by the plus sign, and the number of cells is shown in color. The histogram was smoothed with the Gaussian function (σ = 6 μm). White arrow denotes the orientation of periodicity. Scale bar, (in A3) A3, B3, and C3, 100 μm. A4, Power spectrum of the distribution of EGFP(+) cells in layer V prepared for the orientation of periodicity. The peak wavelength = 28.2 μm. Bar indicates the wavelength range for significance analysis (27.5–30.5 μm). p < 0.0001. B, Analysis of another slice from the same mouse as in A. σ = 6 μm in B3. The peak wavelength in B4 = 29.8 μm. The wavelength range is 27.5–30.5 μm. p = 0.0007. C, Analysis of a slice from another mouse at P35. σ = 6 μm in C3. The peak wavelength in C4 = 29.0 μm. The wavelength range is 27.5–30.5 μm. p = 0.0026. D–F, Autocorrelograms of the positions of EGFP(+) cells in layer V in slices analyzed in A–C. Autocorrelograms were prepared for the orientation of periodicity. Gray vertical lines are placed every 28.2, 29.5, and 29.0 μm, respectively. Gray lateral lines show the highest 2.5%, the highest 15.9%, the median, the lowest 15.9%, and the lowest 2.5% of autocorrelograms calculated for random surrogates. G, H, Identical autocorrelograms as in D and E, shown for longer distance.
