Figure 7.
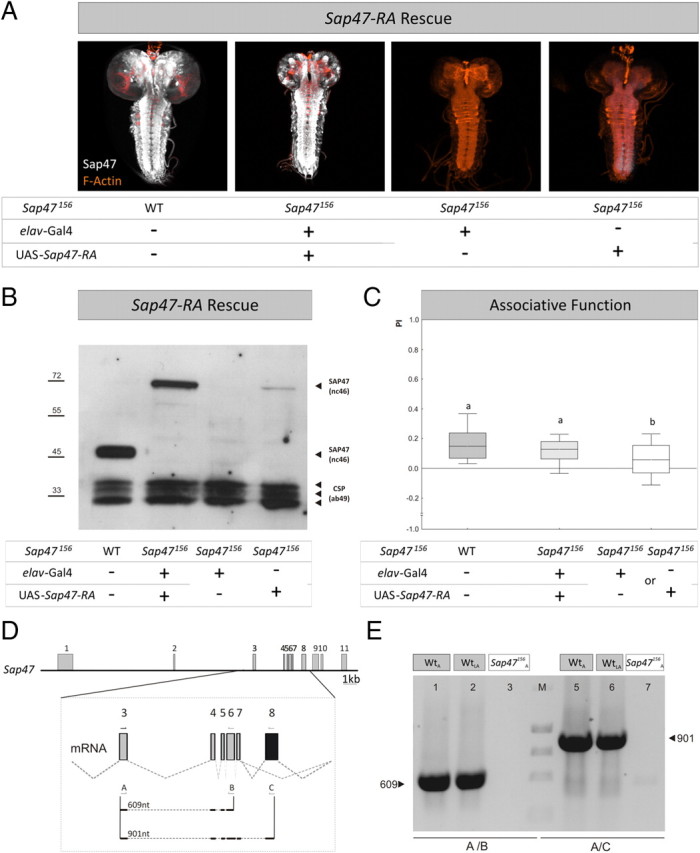
SAP47 full-length rescue. A, Whole mounts. In contrast to WT and rescue larvae, the elav-Gal4; Sap47156 driver-control shows no SAP47 expression, whereas there is a weak leaky expression detectable at the caudal tip of the ventral nerve cord of the UAS-Sap47-RA; Sap47156 effector-control strain. Phalloidin is used to visualize F-actin (orange). B, Western blot. The WT shows the prominent band at 47 kDa, whereas the rescue strain shows the band of the full-length PA isoform of SAP47 of ∼70 kDa, as to be expected from the coding region used for the UAS-Sap47-RA element. No SAP47 signal is detectable in the driver-control, but a weak leaky expression is seen in the UAS-Sap47-RA; Sap47156 effector-control strain, corresponding to the expression seen in the whole mount at the caudal tip of the ventral nerve cord (A). Anti-SAP47: nc46; anti-CSP: ab49 (loading control). C, Expression of the full-length isoform of SAP47 fully rescues associative function. Driver and effector-control are impaired in associative function to ∼50% of control level (WT), using elav-Gal4; Sap47156 as driver- and UAS-Sap47-RA; Sap47156 as effector-control. Both controls perform equally well and are therefore pooled as genetic controls (supplemental Fig. 8C, available at www.jneurosci.org as supplemental material). Larvae expressing the full-length cDNA of SAP47 perform as good as WT. N = 40, 40, 80. Different lettering above plots signifies p < 0.05/2 in Mann–Whitney U tests. The PREF scores underlying all PI values are documented in supplemental Figure 6 (available at www.jneurosci.org as supplemental material). D, Gene structure of SAP47 in WT larvae. Shown is the gene structure of Sap47 with exons and introns in wild type (exons gray, see also Fig. 1A). Eight transcripts are annotated, five longer and 3 shorter transcripts (Flybase: http://flybase.bio.indiana.edu). All of the longer transcripts contain exon 8, whereas this exon is spliced out in all of the short transcripts, indicated by black color in the magnification of the mRNA from exon 3 to exon 8. This situation allows differentiating the long from the short transcripts. The arrows and letters indicate binding sites of primers used for PCR in E and size of the expected PCR products of 609 nt using primer pair A and B and of 901 nt using primer pair A and C. E, PCR. After isolation of total RNA from 100 WT adult flies (WTA), 200 WT third-instar larvae (WTLA), and 100 adult SAP47156 flies (SAP47156A) and producing Sap47 cDNAs, the primer pair A/B generates a 609 nt fragment only in WT. Also using primer pair A/C, a 901 nt fragment is generated in only WT, and notably in both larvae and adults, indicating that long isoforms of SAP47 are expressed. Note that, on the protein level, these longer isoforms are often hard to detect (see Western blot in B).
