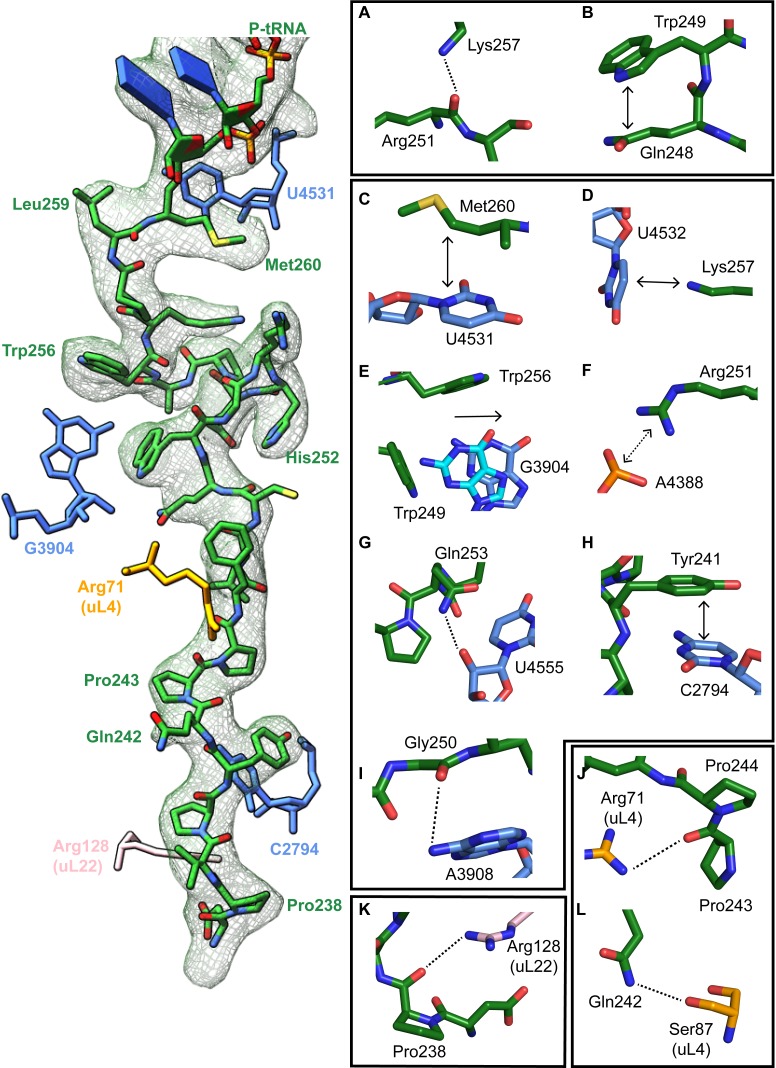
Figure 3. Stabilizing interactions of XBP1u nascent chain with the ribosomal exit tunnel.
On the left shown nascent chain model (green) with density (gray mesh), and some interacting 28S rRNA bases and ribosomal protein residues are shown. Isolated nascent chain density is displayed at contour level of 1.28 σ. (A) Lys257 of XBP1u (green) is at the hydrogen bond making distance internally within XBP1u residue Arg251. (B) Trp249 of the XBP1u stacks internally onto Gln248. (C) Met260 of XBP1u makes a hydrophobic interaction with U4531 of 28S rRNA (blue). (D) Lys257 stacking with the base U4532 (E) Trp256 and Trp249 of XBP1u displace a ribosomal tunnel base G3904 (blue). G3904 conformation with XBP1u is compared with didemnin B treated ribosome (cyan, PDB ID 5LZS). (F) Arg251 of XBP1u makes a salt-bridge interaction with the exit tunnel base A4388. (I, G) Gly250 and Gln253 of XBP1u are in the distance for making hydrogen bond interaction with 28S rRNA bases A3908 and U4555. (H) Tyr241 of XBP1u stacks onto C2794. (J–L) Constriction site protein residues making interaction with XBP1u are shown: uL4 (orange) and uL22 (pink).