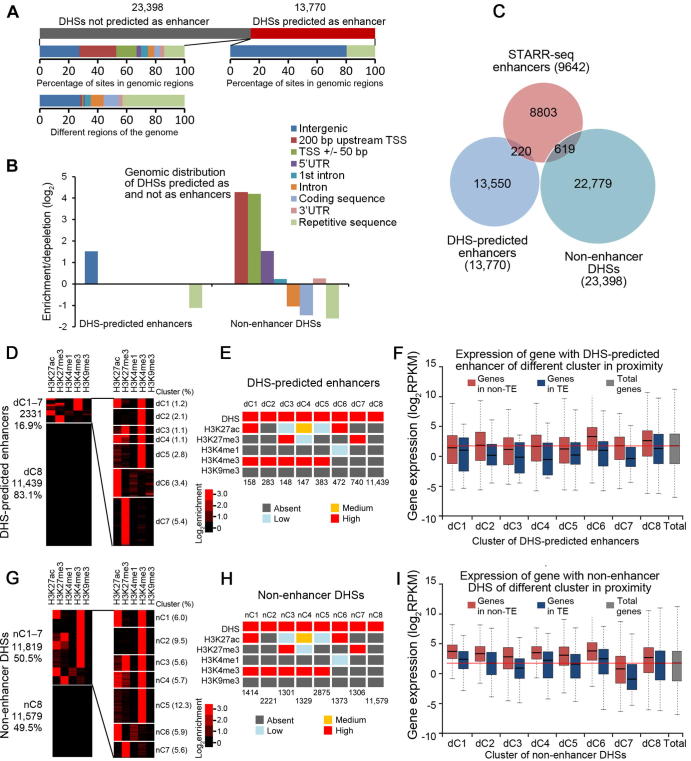
Figure 6.
Predicted enhancers based on DHS location
A. Number of DHS-predicted enhancers (13,770) and non-enhancer DHSs (23,398) and percentage of these sites in different functional genomic regions are shown below. B. Relative enrichment/depletion of DHS-predicted enhancers and non-enhancer DHSs in different genomic regions. C. Overlap analysis among STARR-seq enhancers, DHS-predicted enhancers, and non-enhancer DHSs. D. DHS-predicted enhancers are clustered over the signal of five histone modifications. E. The level of enrichment of epigenetic mark for each cluster of enhancers is shown in different color (see Figure S10B for details). F. Expression of genes with proximal DHS-predicted enhancers in TE (blue) or non-TE (red) regions. G. Non-enhancer DHSs are clustered over the signal of five histone modifications as for STARR-seq enhancers. H. The level of enrichment of epigenetic mark for each cluster of enhancers is shown in different color (see Figure S10C for details). For panels E and H, signal density of clusters is ranked as absent (dark gray), low (light blue), medium (orange), and high (red). The signal density is determined by the percentage of enhancers in a specific cluster carrying the examined epigenetic mark. Number of enhancers in each cluster is shown at the bottom. A cluster is designated to be absent of an epigenetic mark if ≤10% of the elements in that cluster carried the epigenetic mark examined, or designated as low, medium, or high if the percentage of elements in a specific cluster carrying an epigenetic mark examined was between 10%–30%, 30%–60%, or >60%. I. Expression of genes with proximal non-enhancer DHSs in TE (blue) or non-TE (red) regions.