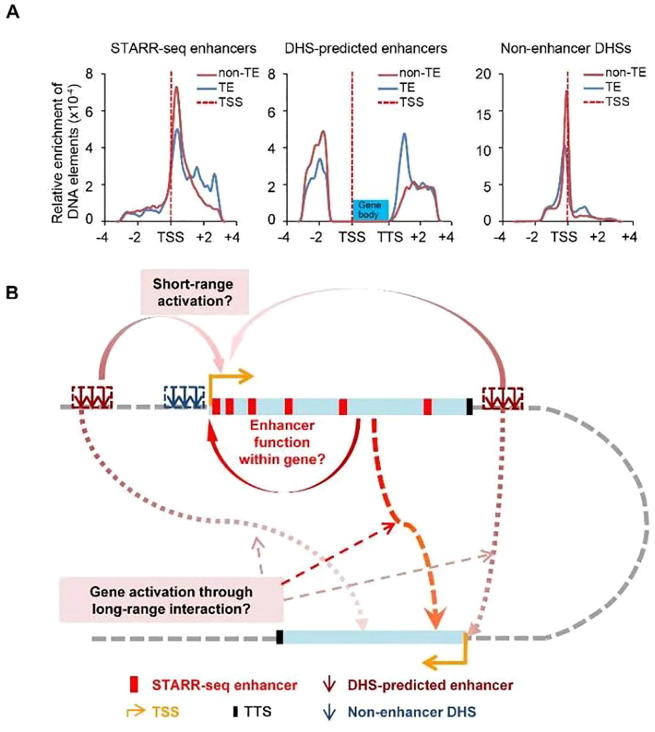
Figure 7.
Distribution of three types of DNA elements and proposed enhancer function model in rice genome
A. Relative enrichment of STARR-seq enhancers, DHS-predicted enhancers, and non-enhancer DHS sites. Red and blue lines show sites in non-TE and TE regions, respectively; the dashed line indicates the location of TSS. B. The proposed model for enhancer distribution and functional mechanisms in rice genome. Gene body is shown as light blue rectangle box. Red boxes indicate STARR-seq enhancers. DHS-predicted enhancers and non-enhancer DHSs are shown as arrows in dark red and dark blue, respectively. Density of STARR-seq enhancers gradually decreases within the gene body from the 5′TSS to the 3′TTS. Genes can be self-regulated by enhancers located within themselves. DHS-predicted enhancers are mostly located not far away from gene bodies. Non-enhancer DHSs are mostly enriched at promoter regions. Enhancers in one gene may activate other genes separated far away linearly but close in three-dimensional space. The start and orientation of transcription are indicated using an orange arrow.