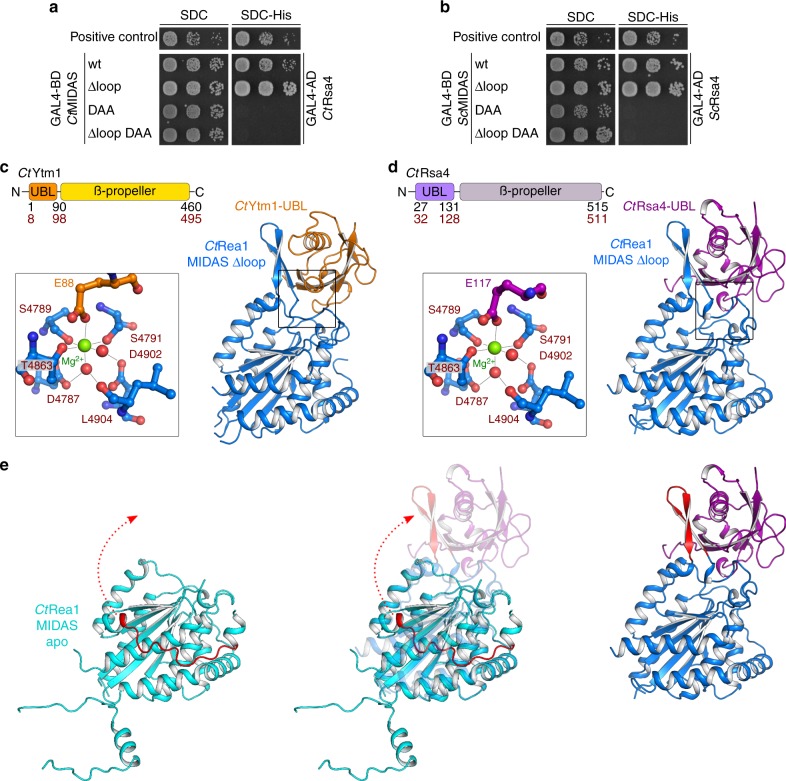
Fig. 2.
The crystal structure of the CtRea1-MIDAS ∆loop with its ligands CtRsa4-UBL and CtYtm1-UBL. a, b Yeast two-hybrid analysis of the interactions between the indicated Rea1-MIDAS constructs and full-length Rsa4 from C. thermophilum (a) and S. cerevisiae (b). The Rea1-MIDAS constructs were fused to an N-terminal GAL4-BD (binding domain) and the Rsa4 constructs were fused to an N-terminal GAL4-AD (activation domain). Plasmids were co-transformed into the PJ69-4A yeast two-hybrid strain and representative transformants were spotted in tenfold serial dilutions on SDC (SDC-Leu-Trp) and SDC-His (SDC-Leu-Trp-His) plates. Cell growth was monitored after incubation for 3 days at 30 °C. Co-transformation of p53 (residues 72–390) fused to the GAL4-BD and SV40 (residues 84–708) fused to the GAL4-AD served as a positive control. c, d Crystal structures of the Rea1-MIDAS domain lacking the protruding element II loop in complex with the UBL domains of Ytm1 (c) and Rsa4 (d) from C. thermophilum. The domain organization of Ytm1 and Rsa4 are shown above the structures. The residue numbers indicate the domain boundaries for S. cerevisiae (black) and C. thermophilum (red). Zoomed-in views of the Mg2+-coordinating residues are shown left of the X-ray structures. The Mg2+ ion is shown in green, the amino acids of the MIDAS consensus motif in red and the glutamate within Ytm1 and Rsa4, which is essential for binding to the MIDAS, in orange (c) and purple (d), respectively. e Comparison of the Rea1-MIDAS structure and the Rea1-MIDAS ∆loop structure in complex with the Rsa4-UBL. The Rea1-specific element III is shown in red, and the rearrangement from its disordered state in the MIDAS apo structure to a β-hairpin in the complex with the Rsa4-UBL is shown