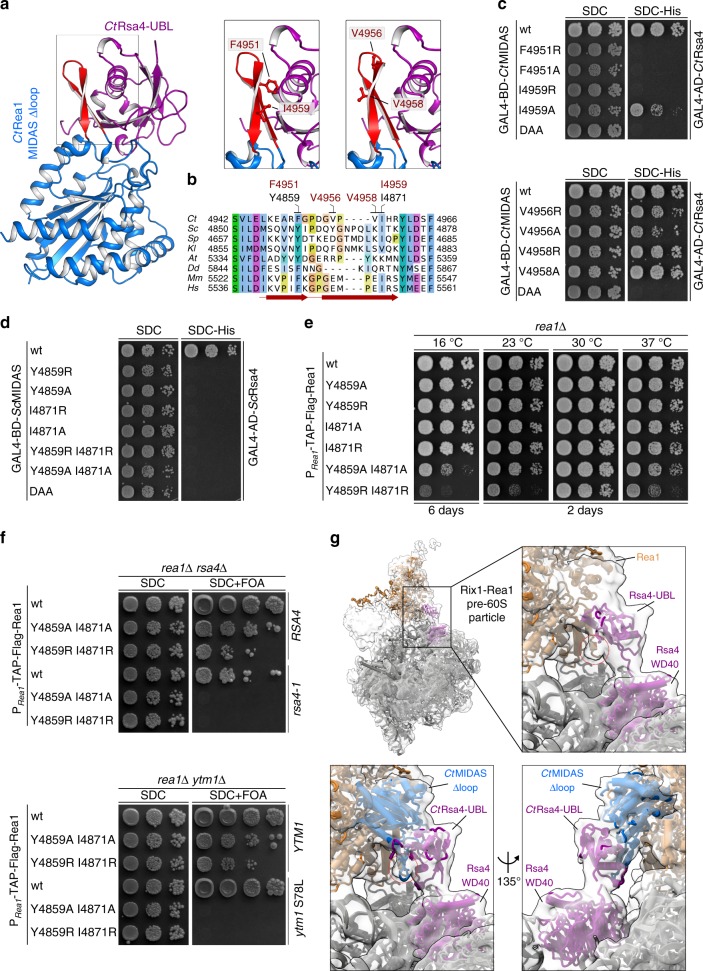
Fig. 3.
The Rea1-MIDAS-specific element III forms a novel interaction site with the UBL domain. a Crystal structure of the CtRea1-MIDAS ∆loop CtRsa4-UBL complex (left panel) and close-up of the interaction between the MIDAS element III and the Rsa4-UBL domain (right panels). The residues selected for mutational analysis are indicated. b Multiple sequence alignment of Rea1 showing the Rea1-MIDAS-specific element III. Sequences of Chaetomium thermophilum (Ct), Saccharomyces cerevisiae (Sc), Schizosaccharomyces pombe (Sp), Kluyveromyces lactis (Kl), Arabidopsis thaliana (At), Dictyostelium discoideum (Dd), Mus musculus (Mm), and Homo sapiens (Hs) were aligned with Clustal Omega and visualized with Jalview. The element III with its two β-sheets are indicated in red, and residues mutated in this study are shown above the alignment (red: C. thermophilum, black: S. cerevisiae). c, d Yeast two-hybrid analysis between the indicated Rea1-MIDAS constructs and full-length Rsa4 from C. thermophilum (c) and S. cerevisiae (d). The MIDAS constructs were fused to an N-terminal GAL4-BD and the Rsa4 constructs to an N-terminal GAL4-AD. The constructs were co-transformed into yeast (PJ69-4A) and cells were spotted on SDC (SDC-Leu-Trp) and SDC-His (SDC-Leu-Trp-His) plates. Cell growth was inspected after 3 days at 30 °C. e The indicated Rea1 constructs were transformed in a rea1∆ shuffle strain. After plasmid shuffling on SDC plates containing 5-FOA, cells were spotted on YPD plates and growth was monitored at the indicated temperatures and times. f The rea1∆ rsa4∆ and rea1∆ ytm1∆ double shuffle strains were transformed with the indicated wild-type and mutant constructs. Transformants were spotted on SDC (SDC-Leu-Trp) and SDC + FOA plates and cell growth at 30 °C was monitored after 3 and 5 days, respectively. g Electron density map of the Rix1–Rea1 particle (EMD-3199) and the models of Rsa4 and Rea1 (PDB ID: 5JCS) in purple and orange, respectively. A density not occupied by the model is highlighted with a red circle (upper close-up). The lower close-up views show the rigid body fit of the Rea1-MIDAS ∆loop–Rsa4-UBL complex from C. thermophilum within the electron density map of the Rix1–Rea1 particle (EMD-3199). The β-hairpin formed by the Rea1-specific element III is highlighted with a red circle