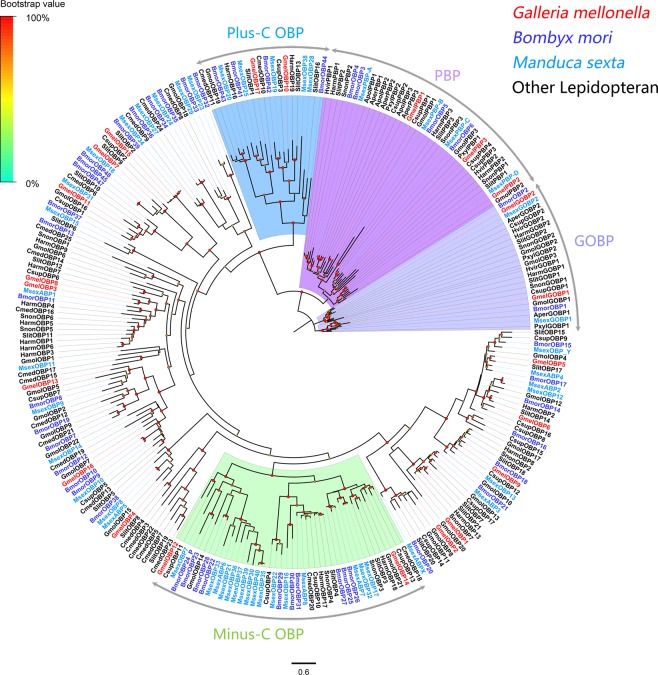
Figure 4.
Maximum-likelihood tree of OBPs. The distance tree was rooted by the conservative GOBP and PBP gene orthologous. Species abbreviations: Gmel, Galleria mellonella; Apol, Antheraea Polyphemus; Aper, Antheraea pernyi; Bmor, Bombyx mori; Cmed, Cnaphalocrocis medinalis; Csup, Chilo suppressalis; Gmol, Grapholita molesta; Pxyl, Plutella xylostella; Slit, Spodoptera litura; Msex, Manduca sexta; Snon, Sesamia nonagrioides. Highlighted clades: pheromone binding proteins, PBPs (purple), general odorant binding proteins, GOBPs (blue), Plus-C OBPs (wathet blue) and Minus-C OBPs (green). Node support (circles at the branch nodes) was estimated using an approximate likelihood ratio test based on the scale indicated at the top left. Bars indicate branch lengths in proportion to amino acid substitutions per site.