Abstract
Aims
Erlotinib is a tyrosine kinase inhibitor used in the treatment of non‐small cell lung cancer highly metabolized by the cytochrome P450 (CYP) 3A. Hence, CYP3A4 activity might be a useful predictor of erlotinib pharmacokinetics in personalized medicine. The effect of erlotinib on CYP3A activity was therefore studied in non‐small cell lung cancer patients.
Methods
The study included 32 patients scheduled for erlotinib monotherapy. CYP3A activity was assessed using quinine as a probe before and during erlotinib treatment. Plasma from blood samples drawn 16 hours post quinine administration were analysed using HPLC with fluorescence detection to determine the quinine/3‐OH‐quinine ratio.
Results
Matched samples, available from 13 patients, showed an induction of CYP3A activity (P = 0.003, Wilcoxon's signed rank test) after 2 months of treatment. The quinine/3‐OH‐quinine ratio decreased from 20.2 (± 13.4) at baseline to 11.0 (± 4.34). Single‐point samples, available from 19 patients, supported the decrease in ratio (P = 0.007, Mann–Whitney U‐test).
Generally, females had a higher CYP3A activity both at baseline and after two months of treatment. Statistical analysis by gender also showed significant increase in CYP3A activity (males, n = 10, P = 0.001, and females, n = 22, P = 0.001).
Conclusions
An induction of CYP3A activity was observed after 2 months of erlotinib treatment which was also seen when subdividing based on gender. It could be important to take this into consideration for patients co‐administering other CYP3A‐metabolizing drugs during erlotinib treatment and also makes it difficult to use baseline CYP3A activity to predict erlotinib pharmacokinetics.
Keywords: CYP3A activity, erlotinib, non‐small cell lung cancer, quinine, Tarceva
What is already known about this subject
Erlotinib (Tarceva®) is a tyrosine kinase inhibitor approved as first‐line treatment in advanced non‐small cell lung cancer patients harbouring EGFR L858R mutation or exon 19 deletions.
Erlotinib is extensively metabolized by CYP3A and known to be a time‐dependent inhibitor of CYP3A activity in vitro.
In general, females have a higher CYP3A activity compared to males.
What this study adds
This study shows that CYP3A activity is induced in non‐small lung cancer patients after 2 months of erlotinib treatment.
The induction of CYP3A activity occurs in both genders despite the observed gender differences in CYP3A activity.
1. INTRODUCTION
Erlotinib (Tarceva®) is an epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor (TKI) that is used in the treatment of advanced non‐small cell lung cancer (NSCLC). It is used as first‐line treatment in patients harbouring a sensitizing mutation in the tyrosine kinase domain such as exon 19 deletion or L858R.
Erlotinib is predominantly eliminated through metabolism by the liver and only 2% is eliminated unchanged.1 The main contributors to the metabolism are the cytochrome P450 (CYP) enzymes, CYP3A4, CYP3A5 and CYP1A1.2 CYP3A4 and CYP3A5 primarily convert erlotinib into the main metabolite OSI‐420.
Toxicities, such as skin rash and diarrhoea, are common in erlotinib treatment and around 75% of the patients experience rash.3 Severe skin toxicities lead to reduced dose or even ceased treatment. A recent study has shown a correlation between erlotinib metabolic ratio (erlotinib/OSI‐420) and skin toxicities, suggesting that metabolism of erlotinib could be of importance for adverse drug reactions.4
Several studies have shown that erlotinib is as a time‐dependent inhibitor of CYP3A in vitro.5, 6, 7 In vivo, CYP3A activity has been studied twice after 8 days and 30 days of erlotinib treatment but did not significantly demonstrate any change in CYP3A activity using midazolam clearance and erythromycin breath test,5 and urinary 6β‐hyroxycortisol as CYP3A activity probes, respectively.8 However, a trend towards increased CYPA3 activity was observed in both studies.
The CYP3A activity probe quinine, used in this project, is hydroxylated by CYP3A into 3‐hydroxyquinine (3‐OH‐quinine). Quinine is metabolized by CYP3A4 and CYP3A5 and has been shown to be a reliable biomarker for CYP3A activity measurements.9, 10
In order to explore whether the CYP3A activity assessed before treatment start can be used as a predictive biomarker for erlotinib pharmacokinetics, the aim of this project was to study the long‐term effect of erlotinib on CYP3A activity in NSCLC patients by analysing CYP3A activity before and after 2 months of erlotinib treatment using quinine as a probe. This could provide useful information in predicting the metabolic capacity and aid in individualization of the treatment strategy before initiation of erlotinib treatment.
2. MATERIAL AND METHODS
2.1. Clinical study
In total, 32 patients diagnosed with NSCLC and scheduled for erlotinib treatment were consecutively included in the study at five centres in Sweden. Exclusion criteria were known allergy to quinine, diagnosed with myasthenia gravis or co‐treatment with CYP3A4 inhibitors such as ketoconazole, itraconazole, erythromycin, clarithromycin, telithromycin or HIV‐protease inhibitors. The patients were administered 100–150 mg erlotinib daily for at least 2 months. All patients provided written informed consent prior to enrolment in accordance with the Helsinki Declaration and the study was approved by the regional ethical board in Linköping (DNR: 2014‐147/32, 2015‐74/32, 2016‐13/32).
2.2. CYP3A phenotyping
CYP3A activity in vivo was determined by measuring hydroxylation of quinine into 3‐OH‐quinine at baseline and after 2 months of treatment. Heparin blood samples were drawn 16 hours (± 2 hours) after administration of a tablet of quinine (250 mg, Recip AB, Solna, Sweden), a validated biomarker for monitoring CYP3A activity.9 The blood samples were centrifuged at 3000g for 10 minutes at room temperature, and the plasma was thereafter transferred to a tube and stored at −20°C until analysis.
2.3. Chemicals and reference substances
Reference substances of quinine and (−)‐(3S)‐3‐hydroxyquinine were purchased from Toronto Research Chemicals (North York, Canada). Acetic acid and ammonium acetate (AmAc), of analytical grade or higher, were purchased from Sigma Aldrich (Stockholm, Sweden). Methanol (MeOH) and acetonitrile (ACN) of HPLC grade were obtained from Fischer Scientific (Göteborg, Sweden).
2.4. Sample preparation
Thawed plasma samples were prepared by precipitating 100 μL of plasma with 200 μL of cold methanol. Samples were vortexed and centrifuged at 3000g for 10 minutes at 4°C. The supernatant was transferred to a vial for analysis.
2.5. Chromatographic conditions
The method for determining the 3‐OH‐quinine and quinine concentration ratio as a measure of CYP3A activity has been described previously.11, 12 Briefly, the samples were analysed using a high‐performance liquid chromatography (HPLC) system (Waters 2695 alliance separation module) with fluorescence detection (Waters 2475 multi λ fluorescence detector). The fluorescence excitation and emission wavelengths used were 350 nm and 450 nm, respectively. The separation took place on an Xbridge C18 column (3.0 × 150 mm, 3.5 μm, Waters) at 60°C. The mobile phase consisted of 0.1 M acetate buffer pH 4 (0.088 M acetic acid, 0.012 M AmAc) and ACN with a flow rate of 0.8 mL/min. The gradient of the 12.5‐minute analysis was 10–14% ACN at 0–5 minutes, 14–26% ACN at 5–9.4 minutes and back to 10% ACN for 9.4–12.5 minutes.
2.6. Calibration and quality control
Six levels of calibration standards for quinine and 3‐OH‐quinine were prepared in heparin drug‐free plasma at 100–10 000 nM (100, 250, 600, 1500, 4000 and 10 000 nM) and 10–2000 nM (10, 30, 80, 250, 700 and 2000 nM), respectively. The lower quantification limits were 100 nM for quinine and 10 nM for 3‐OH‐quinine. Quality control samples were prepared at low, medium and high concentration levels for quinine (150, 1000 and 7500 nM) and 3‐OH‐quinine (20, 150 and 1500 nM), respectively.
2.7. Statistics
All statistical analyses were performed using IBM SPSS Statistics, version 23 (IBM, Armonk, NY, USA). Matched samples were analysed with Wilcoxon's signed rank test, otherwise the Mann–Whitney U‐test was used.
3. RESULTS
3.1. Patient population
All included patients were diagnosed with adenocarcinoma and the majority were females (69%). All patient characteristics are listed in Table 1. No patients were identified to co‐administer any CYP3A or ABCB1 modulating substances except for two patients that were administering a weak CYP3A inhibitor.
Table 1.
Patient characteristics (n = 32)
| Patient characteristics | n (%) |
|---|---|
| Age | |
| Median age | 67 |
| Range | (51–87) |
| Gender | |
| Female | 22 (68.8) |
| Male | 10 (31.3) |
| Clinical stage | |
| Stage III | 3 (9.4) |
| Stage IV | 29 (90.6) |
| Smoking history | |
| Never | 16 (50.0) |
| Former | 13 (40.6) |
| Current | 3 (9.4) |
3.2. Analysis of quinine/3‐OH‐quinine ratio
In the analytical method for quantification of 3‐OH‐quinine and quinine, the substances eluted after 4.9 and 9.1 minutes, respectively. The performance of the method was monitored in each batch (n = 7). Precision and accuracy for quinine and 3‐OH‐quinine at low‐, medium‐ and high‐quality control levels were <3% and between 93% and 101%, respectively.
3.3. CYP3A metabolic activity
Matched samples, drawn at baseline and after 2 months of treatment were available for 13 of the 32 enrolled patients. The quinine/3‐OH‐ratio in these 13 patients was significantly lower after 2 months of erlotinib treatment compared to before start of erlotinib treatment (P = 0.003), which is consistent with a higher CYP3A4 activity at 2 months (Figure 1A and Table 2). On average, a 2‐fold induction was observed in the matched samples, 2.7‐fold in males and 1.6‐fold in females.
Figure 1.
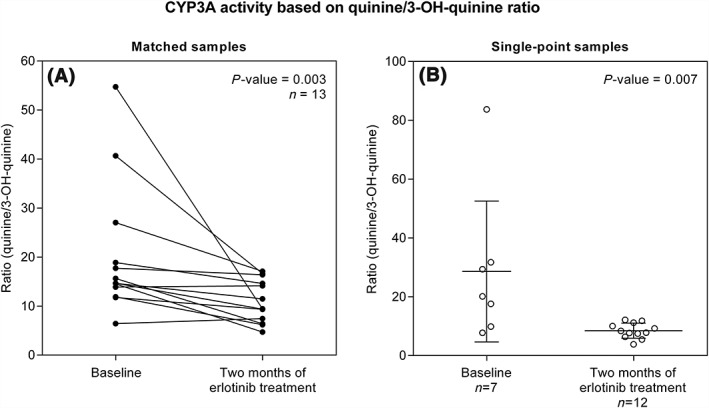
Ratio of quinine/3‐OH‐quinine as a measure of CYP3A activity before start of treatment (baseline) and after 2 months of erlotinib monotherapy, in (A) matched samples (n = 13) and (B) single‐point samples (n = 7 at baseline and n = 12 at 2 months of treatment). The statistically significant decrease in quinine/3‐OH‐quinine ratio (increased CYP3A activity) observed in matched samples (P = 0.003) was supported in single‐point samples (P = 0.007). Error bars in (B) illustrates mean with 95% CI
Table 2.
Summary of quinine/3‐OH‐quinine ratio
| Baseline | 95% CI | 2 months of erlotinib treatment | P‐value | Average level of induction | |||
|---|---|---|---|---|---|---|---|
| n | Average ratio (± SD) | Average ratio (± SD) | 95% CI | ||||
| Matched samples | 13 | 20.2(± 13.4) | 12.1–28.3 | 11.0 (± 4.34) | 8.41–13.7 | 0.003 | 2.0 |
| Males | 4 | 35.1 (± 16.2) | 9.33–60.8 | 14.9 (± 3.65) | 9.09–20.7 | 0.068 | 2.7 |
| Females | 9 | 13.6 (± 3.41) | 11.0–16.2 | 9.31 (± 3.52) | 6.61–12.0 | 0.021 | 1.6 |
Single point measurements, available from 19 patients, 7 patients with baseline measurements and 12 patients with samples from 2 months of treatment, were used to validate the findings from the 13 matched patients. Ratios consistent with an inductive effect were also observed in this group (P = 0.007) (Figure 1B). The average ratio was 28.6 (± 25.9) at baseline and 8.46 (± 2.56) after 2 months of erlotinib treatment.
Gender was shown to have an impact on the study when analysing all available patients: 20 patients at baseline and 25 patients after two months of treatment (n = 32) (Figure 2). The variation in quinine/3‐OH‐quinine ratio in all samples showed a 13‐fold and 5‐fold variation in the metabolic ratio at baseline and after 2 months of treatment. The 13‐fold variation was considerably higher than previously described.9, 13 When examining each time point by gender, males showed a 3‐fold variation at baseline and a 2‐fold variation after 2 months of treatment (Figure 2B). Females showed a 13‐fold variation at baseline and a 4‐fold variation after 2 months of treatment (Figure 2C). The 13‐fold variation in females at baseline decreases to a 3‐fold variation when ignoring one outlier with unusually high metabolic ratio (Figure 2C). The quinine/3‐OH‐quinine ratio was thereafter shown to be lower in females compared to males at each time point, baseline and after 2 months of treatment, in all samples (P = 0.002 and P = 0.003), indicating overall higher CYP3A activity in females (Figure 2A). Separate analyses of females and males in all samples showed statistical differences in the CYP3A activity independent of gender (P = 0.001) (see Figures 2B and 2C and Table 2).
Figure 2.
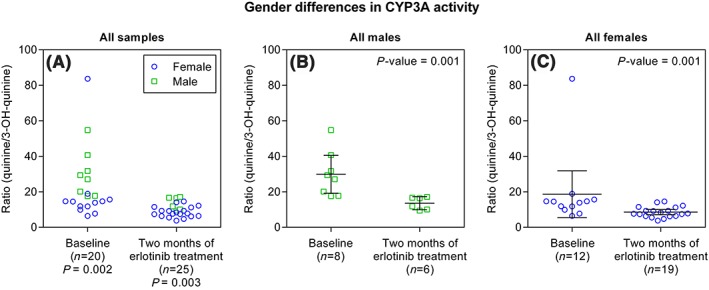
Illustration of gender differences in CYP3A activity based on the entire study population where (A) females show lower quinine/3‐OH‐quinine ratio compared to males at baseline and after 2 months of erlotinib treatment. When all patients are subdivided based on gender, into (B) males and (C) females, the statistically significant decrease in quinine/3‐OH‐quinine ratios between samples obtained at baseline and at 2 months of treatments is maintained
4. DISCUSSION
In this project, we studied CYP3A activity in NSCLC patients before and after 2 months of erlotinib treatment. We found that the quinine/3‐OH‐quinine ratio significantly decreases in patients after 2 months of erlotinib treatment, which indicates that erlotinib induces CYP3A activity.
The number of patients available with matched samples from baseline and 2 months of erlotinib treatment was 13 in total. The remaining 19 patients provided samples from only one time point. To utilize the data from all patients, the single‐point samples were assigned to be a validation group. The groups were thereafter analysed separately with paired and unpaired statistics. The significant results obtained by the single‐point samples therefore strengthen the observation of an inductive effect of CYP3A in the matched samples.
CYP3A activity has been studied previously in patients after 1 week and 30 days of erlotinib administration.5, 8 Induction could not be statistically determined even though the results pointed in the direction of increased CYP3A activity. However, in the present study, we identified a 2‐fold induction of CYP3A activity by erlotinib when the CYP3A activity was assessed after 2 months of erlotinib treatment. The differences from previous studies were the choice of probe and time of erlotinib treatment.
The probe used for CYP3A activity measurements in this study was quinine. It has been shown to be a stable biomarker for CYP3A activity and has been suggested to represent the hepatic CYP3A metabolism.9 It is easy to use as only one sample is required to assess the CYP3A metabolic activity. One of the advantages with quinine as a CYP3A activity probe is that the patient can administer the drug at home 16 hours before sampling. That can also be a possible source of error because it is difficult to control for compliance. Quinine, as well as erlotinib, is a substrate of P‐glycoprotein (P‐gp, MDR1, ABCB1).14, 15 A study in mice showed that the quinine/3‐OH‐quinine ratio is similar in mice with and without P‐gp,15 suggesting that a possible inhibition of P‐gp would not influence the quinine metabolic ratio substantially.
There are several other probes available to assess CYP3A activity. Two common probes are the erythromycin breath test and midazolam clearance.16, 17 The erythromycin breath test includes intravenous administration of radioactively labelled erythromycin. Midazolam, on the other hand, is administered the same way as quinine, in tablet form. Unlike quinine, midazolam clearance must be monitored over time and several samples must be drawn but has the advantage that it measures both intestinal and hepatic CYP3A.18 Other CYP3A probes that also can be measured are the endogenous 4‐β‐hydroxycholesterol from serum and 6‐β‐hydroxycholesterol from urine.19, 20 One possible drawback with 4‐β‐hydroxycholesterol is a long half‐life of the substance and therefore rapid changes in CYP3A activity cannot be detected.21 The different probes have been compared and it has been shown that they perform equally well when it comes to detecting CYP3A induction.20
Gender was also shown to have an influence on the quinine/3‐OH‐quinine ratio in this study. These results were partly anticipated as CYP3A activity is known to be higher in women as previously shown.19, 22
Our study shows a 2‐fold induction of CYP3A activity during erlotinib treatment, which might affect the cancer treatment since erlotinib is extensively metabolized by both CYP3A4 and CYP3A5.2 It could, for instance, lead to reduced plasma concentrations and decreased anti‐tumour effect. Patients on erlotinib treatment can also be co‐administering drugs that can be subjected to CYP3A metabolism at the same time. In that case, it is important to be aware of the inducing effect erlotinib can have on CYP3A. However, the clinical significance is uncertain as the effect was only evaluated based on the metabolic ratio of quinine and the effect should be confirmed using a second CYP3A phenotyping approach.
Today, patients start erlotinib treatment at the same dose, a 150 mg daily tablet. Toxicities in erlotinib treatment, such as skin rash and diarrhoea, are common and usually appear after approximately 1–2 weeks of treatment.23 Some patients experience severe toxicities while others do not. Steady‐state plasma concentrations are also known to display a large variation in patients treated with the same dose.24 Recently erlotinib metabolic ratio (erlotinib/OSI‐420) was correlated to skin rash, response and overall survival.4 Due to the fixed dose, the variation of toxicity and changes in CYP3A activity, there might be an opportunity to individualize the treatment in patients by further studying CYP3A activity in patients along with plasma concentration during erlotinib treatment. Another implication of this study might be that, as erlotinib induces its own metabolizing enzyme, the metabolic capacity increases with time, indicating a possible need for dose modifications and therapeutic drug monitoring.
In conclusion, we have shown that the CYP3A activity is induced in patients after 2 months of erlotinib treatment compared to the activity before treatment start. This could be of importance to take into consideration when trying to individualize treatment with erlotinib and if patients are co‐prescribed drugs metabolized by CYP3A during erlotinib treatment.
COMPETING INTERESTS
The authors declare no competing interests.
CONTRIBUTORS
H.G. and S.V. conceived and designed the study. A.S., S.V. and H.G. developed the methodology used in the study and interpreted the data. A.V., N.H., M.K., E.B., H.K., B.B., and H.G. acquired and managed patients, data and/or provided facilities. A.S. conducted the statistical analysis in the study. All authors held administrative roles for reporting and organizing data as well as read and approved the final manuscript.
ACKNOWLEDGEMENTS
We greatly acknowledge participating patients in this study and the hospital personnel for assistance in collecting samples. We are also grateful to the Medical Research Council of Southeast Sweden, Swedish Cancer Society, the Swedish Research Council and Linköping University for funding this study.
Svedberg A, Vikingsson S, Vikström A, et al. Erlotinib treatment induces cytochrome P450 3A activity in non‐small cell lung cancer patients. Br J Clin Pharmacol. 2019;85:1704–1709. 10.1111/bcp.13953
PI Statement: Anders Vikström was the principal investigator in this study.
REFERENCES
- 1. Ling J, Johnson KA, Miao Z, et al. Metabolism and excretion of erlotinib, a small molecule inhibitor of epidermal growth factor receptor tyrosine kinase, in healthy male volunteers. Drug Metab Dispos. 2006;34(3):420‐426. [DOI] [PubMed] [Google Scholar]
- 2. Li J, Zhao M, He P, Hidalgo M, Baker SD. Differential metabolism of gefitinib and erlotinib by human cytochrome P450 enzymes. Clin Cancer Res. 2007;13(12):3731‐3737. [DOI] [PubMed] [Google Scholar]
- 3. Perez‐Soler R, Chachoua A, Hammond LA, et al. Determinants of tumor response and survival with erlotinib in patients with non‐small‐cell lung cancer. J Clin Oncol. 2004;22(16):3238‐3247. [DOI] [PubMed] [Google Scholar]
- 4. Steffens M, Paul T, Hichert V, et al. Dosing to rash? The role of erlotinib metabolic ratio from patient serum in the search of predictive biomarkers for EGFR inhibitor‐mediated skin rash. Eur J Cancer. 2016;55:131‐139. [DOI] [PubMed] [Google Scholar]
- 5. Calvert H, Twelves C, Ranson M, et al. Effect of erlotinib on CYP3A activity, evaluated in vitro and by dual probes in patients with cancer. Anticancer Drugs. 2014;25(7):832‐840. [DOI] [PubMed] [Google Scholar]
- 6. Dong PP, Fang ZZ, Zhang YY, et al. Substrate‐dependent modulation of the catalytic activity of CYP3A by erlotinib. Acta Pharmacol Sin. 2011;32(3):399‐407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kenny JR, Mukadam S, Zhang C, et al. Drug–drug interaction potential of marketed oncology drugs: In vitro assessment of time‐dependent cytochrome P450 inhibition, reactive metabolite formation and drug–drug interaction prediction. Pharm Res. 2012;29(7):1960‐1976. [DOI] [PubMed] [Google Scholar]
- 8. Tiseo M, Andreoli R, Gelsomino F, et al. Correlation between erlotinib pharmacokinetics, cutaneous toxicity and clinical outcomes in patients with advanced non‐small cell lung cancer (NSCLC). Lung Cancer. 2014;83(2):265‐271. [DOI] [PubMed] [Google Scholar]
- 9. Mirghani RA, Ericsson Ö, Tybring G, Gustafsson LL, Bertilsson L. Quinine 3‐hydroxylation as a biomarker reaction for the activity of CYP3A4 in man. Eur J Clin Pharmacol. 2003;59(1):23‐28. [DOI] [PubMed] [Google Scholar]
- 10. Allqvist A, Miura J, Bertilsson L, Mirghani RA. Inhibition of CYP3A4 and CYP3A5 catalyzed metabolism of alprazolam and quinine by ketoconazole as racemate and four different enantiomers. Eur J Clin Pharmacol. 2007;63(2):173‐179. [DOI] [PubMed] [Google Scholar]
- 11. Skoglund K, Richter J, Olsson‐Strömberg U, et al. In vivo cytochrome P450 3A isoenzyme activity and pharmacokinetics of imatinib in relation to therapeutic outcome in patients with chronic myeloid leukemia. Ther Drug Monit. 2016;38(2):230‐238. [DOI] [PubMed] [Google Scholar]
- 12. Mirghani RA, Ericsson Ö, Cook J, Yu P, Gustafsson LL. Simultaneous determination of quinine and four metabolites in plasma and urine by high‐performance liquid chromatography. J Chromatogr B Biomed Sci Appl. 2001;754(1):57‐64. [DOI] [PubMed] [Google Scholar]
- 13. Damkier P, Brosen K. Quinidine as a probe for CYP3A4 activity: Intrasubject variability and lack of correlation with probe‐based assays for CYP1A2, CYP2C9, CYP2C19, and CYP2D6. Clin Pharmacol Ther. 2000;68(2):199‐209. [DOI] [PubMed] [Google Scholar]
- 14. Marchetti S, de Vries NA, Buckle T, et al. Effect of the ATP‐binding cassette drug transporters ABCB1, ABCG2, and ABCC2 on erlotinib hydrochloride (Tarceva) disposition in in vitro and in vivo pharmacokinetic studies employing Bcrp1−/−/Mdr1a/1b−/− (triple‐knockout) and wild‐type mice. Mol Cancer Ther. 2008;7(8):2280‐2287. [DOI] [PubMed] [Google Scholar]
- 15. Pussard E, Merzouk M, Barennes H. Increased uptake of quinine into the brain by inhibition of P‐glycoprotein. Eur J Pharm Sci. 2007;32(2):123‐127. [DOI] [PubMed] [Google Scholar]
- 16. Watkins PB, Murray SA, Winkelman LG, Heuman DM, Wrighton SA, Guzelian PS. Erythromycin breath test as an assay of glucocorticoid‐inducible liver cytochromes P‐450: Studies in rats and patients. J Clin Investig. 1989;83(2):688‐697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kronbach T, Mathys D, Umeno M, Gonzalez FJ, Meyer UA. Oxidation of midazolam and triazolam by human liver cytochrome P450IIIA4. Mol Pharmacol. 1989;36(1):89‐96. [PubMed] [Google Scholar]
- 18. Gorski JC, Jones DR, Haehner‐Daniels BD, Hamman MA, O'Mara EM, Hall SD. The contribution of intestinal and hepatic CYP3A to the interaction between midazolam and clarithromycin. Clin Pharmacol Ther. 1998;64(2):133‐143. [DOI] [PubMed] [Google Scholar]
- 19. Diczfalusy U, Miura J, Roh HK, et al. 4β‐hydroxycholesterol is a new endogenous CYP3A marker: Relationship to CYP3A5 genotype, quinine 3‐hydroxylation and sex in Koreans, Swedes and Tanzanians. Pharmacogenet Genomics. 2008;18(3):201‐208. [DOI] [PubMed] [Google Scholar]
- 20. Björkhem‐Bergman L, Bäckström T, Nylén H, et al. Quinine compared to 4β‐hydroxycholesterol and midazolam as markers for CYP3A induction by rifampicin. Drug Metab Pharmacokinet. 2014;29(4):352‐355. [DOI] [PubMed] [Google Scholar]
- 21. Diczfalusy U, Kanebratt KP, Bredberg E, Andersson TB, Böttiger Y, Bertilsson L. 4β‐hydroxycholesterol as an endogenous marker for CYP3A4/5 activity: Stability and half‐life of elimination after induction with rifampicin. Br J Clin Pharmacol. 2009;67(1):38‐43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hunt CM, Westerkam WR, Stave GM. Effect of age and gender on the activity of human hepatic CYP3A. Biochem Pharmacol. 1992;44(2):275‐283. [DOI] [PubMed] [Google Scholar]
- 23. Wacker B, Nagrani T, Weinberg J, Witt K, Clark G, Cagnoni PJ. Correlation between development of rash and efficacy in patients treated with the epidermal growth factor receptor tyrosine kinase inhibitor erlotinib in two large phase III studies. Clin Cancer Res. 2007;13(13):3913‐3921. [DOI] [PubMed] [Google Scholar]
- 24. Hidalgo M, Siu LL, Nemunaitis J, et al. Phase I and pharmacologic study of OSI‐774, an epidermal growth factor receptor tyrosine kinase inhibitor, in patients with advanced solid malignancies. J Clin Oncol. 2001;19(13):3267‐3279. [DOI] [PubMed] [Google Scholar]


