Figure 3. Genetic loss of EFG1 underlies the white-to-gray transition in vitro.
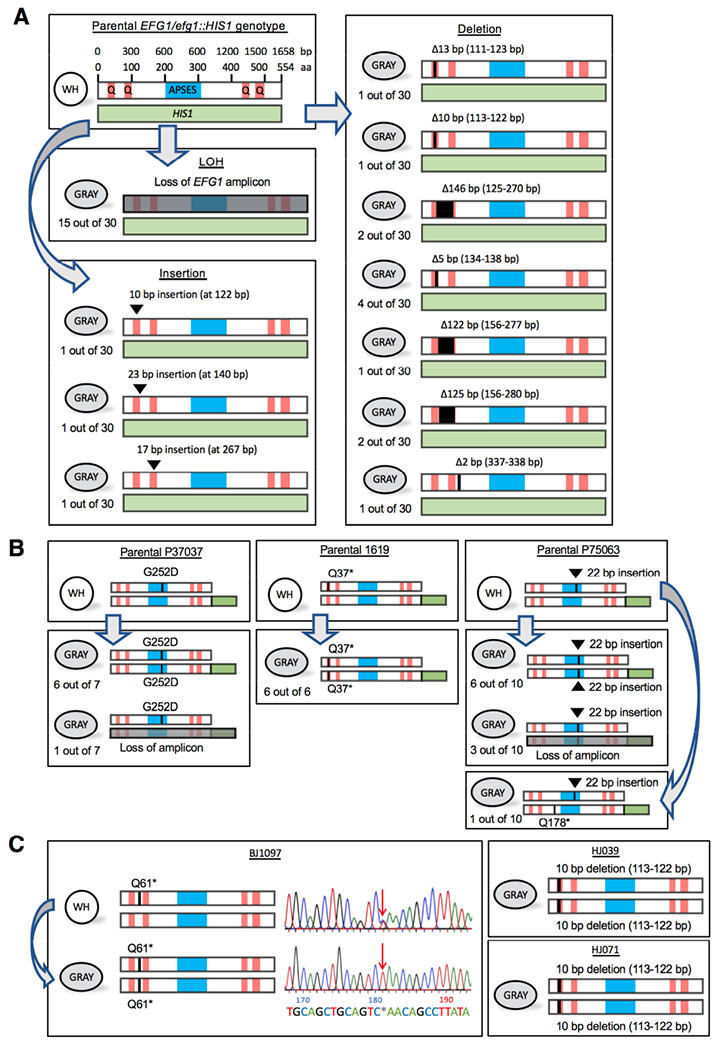
(A) Schematics show EFG1 mutations from white-to-gray switching. APSES, DNA binding domain. Q, polyglutamine tracts. The parental SC5314 strain is heterozygous for EFG1 (one allele replaced with HIS1) and mutational patterns are shown for 30 independent white-to-gray transitions.
(B) Schematics depicting EFG1 mutations in three clinical strains (P37037, 1619, and P75063) upon switching to the gray state. The intact EFG1 allele was tagged with GFP (green box) in the parental strain. Independent switching events are shown.
(C) Schematic of EFG1 mutations present in three clinical strains (BJ1097, HJ039, and HJ071) in which the gray state was first identified. For BJ1097, the chromatogram shows Sanger sequencing of EFG1 in white and gray cells. Arrow indicates a mutation (C181T) causing a premature stop codon (Q61*) that is heterozygous in white cells but homozygous in gray cells.
See also Figure S4 and S5.
