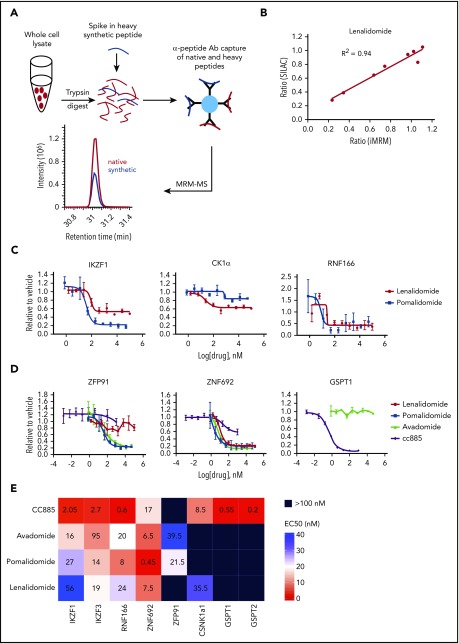
Figure 1.
Substrate degradation differs between drugs. (A) Schematic representation of the sample processing and analysis for the iMRM assay. Native peptide present in the sample and synthetic heavy peptide spiked into the sample are indicated by red and blue color, respectively. Liquid chromatography (LC)–MRM-MS trace is shown for 1 of the peptides, however, all 22 peptides were monitored in a single LC-MRM-MS method. (B) Plot of the ratio of substrate level in 1 µM lenalidomide over vehicle as measured in the iMRM assay vs SILAC MS. Line represents the linear regression (R2 = 0.94). (C-D) MM1S cells were treated with serial dilutions of lenalidomide (red), pomalidomide (blue), avadomide (green), CC-885 (purple), or vehicle for 6 hours and then substrate degradation was measured using the iMRM assay. Protein levels were normalized to housekeeping proteins (2 β-actin peptides and 1 GAPDH peptide) and the level in the vehicle-treated sample was normalized to 1. For each protein, data for a single peptide are shown. Data are mean ± standard error of the mean (SEM) (n = 3 replicates). Curves represent the logistic regression. (E) Logistic regression was used to calculated the EC50 for each drug and protein pairing. The value represents the mean EC50 value for all evaluable peptides (n = 1-2) for each protein. Ab, antibody.