Fig. 1.
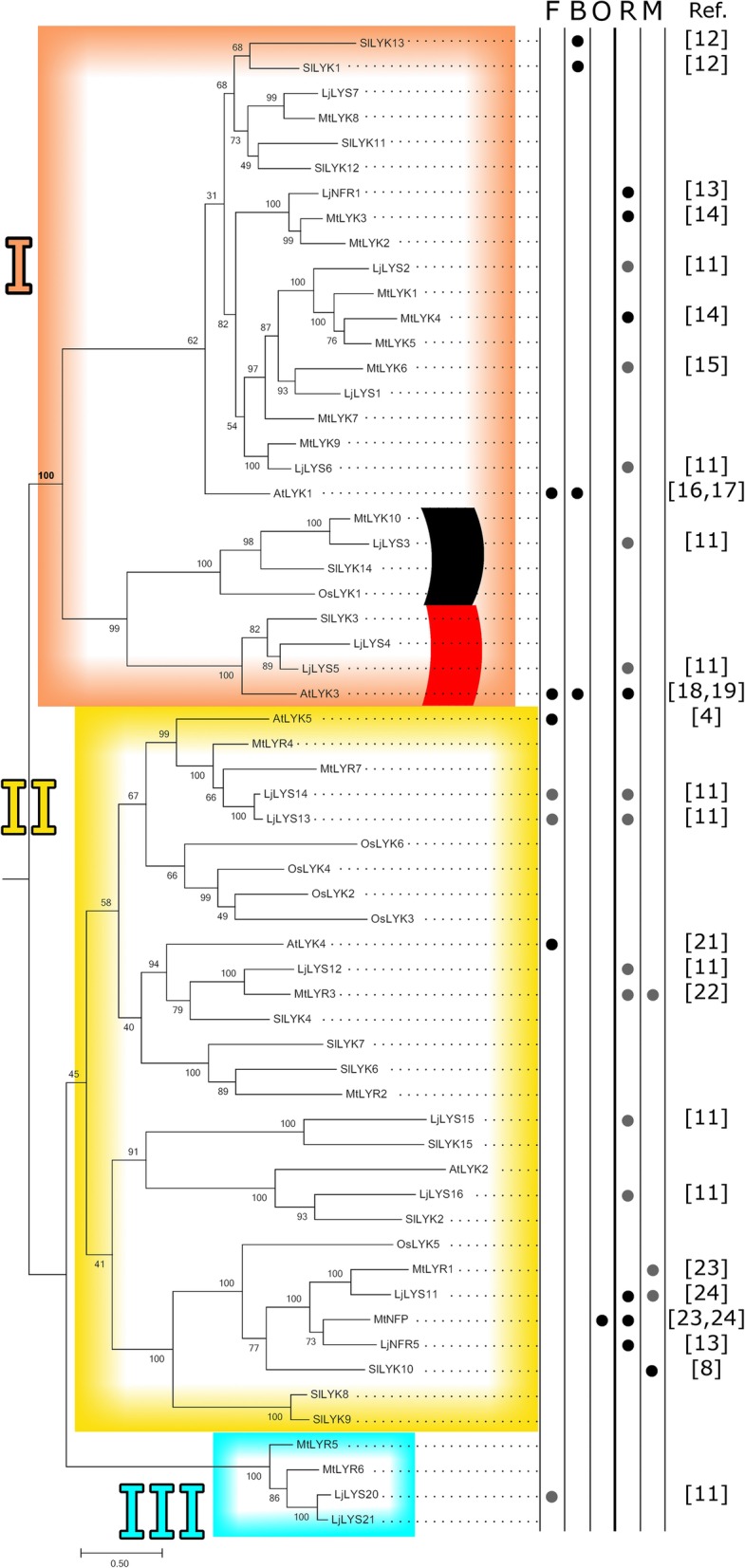
LysM-RLK phylogeny and functions. Maximum likelihood phylogeny and functions of canonical LysM-RLKs from Solanum lycopersicum (Sl), Arabidopsis thaliana (At), Lotus japonicus (Lj), Oryza sativa (Os), and Medicago truncatula (Mt). The phylogeny and 500 bootstrap replicates were inferred using RAxML under the WAG model with empirical frequencies and seed values of 100. The species of gene origin is given by the first two letters of the name given on the phylogeny. The phylogeny was rooted using the method of Minimal Ancestor Deviation [27]. The scale bar indicates amino acid substitutions per site. Gene functions are indicated: defense against fungi (F), bacteria (B), and oomycetes (O) and symbiosis with rhizobia (R) and mycorrhiza (M). LysM-RLKs form three clades. Red and black arcs indicate groups of proteins with distinct LysM domain sequences. Functions verified by mutation phenotypes are indicated by black circles. Functions inferred by differential expression are indicated by gray circles. Citations for sources of functional information are shown in brackets
