Fig. 5.
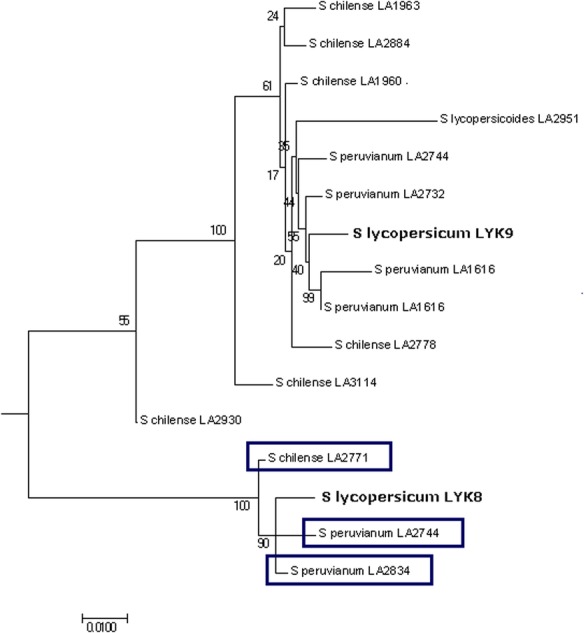
Phylogeny of wild tomato SlLYK8 and SlLYK9 orthologs with intact kinases. This phylogeny includes BLAST hits for Solanum lycopersicum LysM-RLKs SlLYK8 and SlLYK9 which extend past the point of SlLYK8 truncation. The phylogeny and 500 bootstrap replicates were inferred using RAxML under the GTRGAMMA model and seed values of 100 and rooted using the method of Minimal Ancestor Deviation [27]. The scale bar indicates nucleotide substitutions per site. Three sequences from Solanum peruvianum and Solanum chilense with intact kinase domains share more recent ancestry with SlLYK8 than with SlLYK9 and show the greatest percent identity with SlLYK8
