Figure 2.
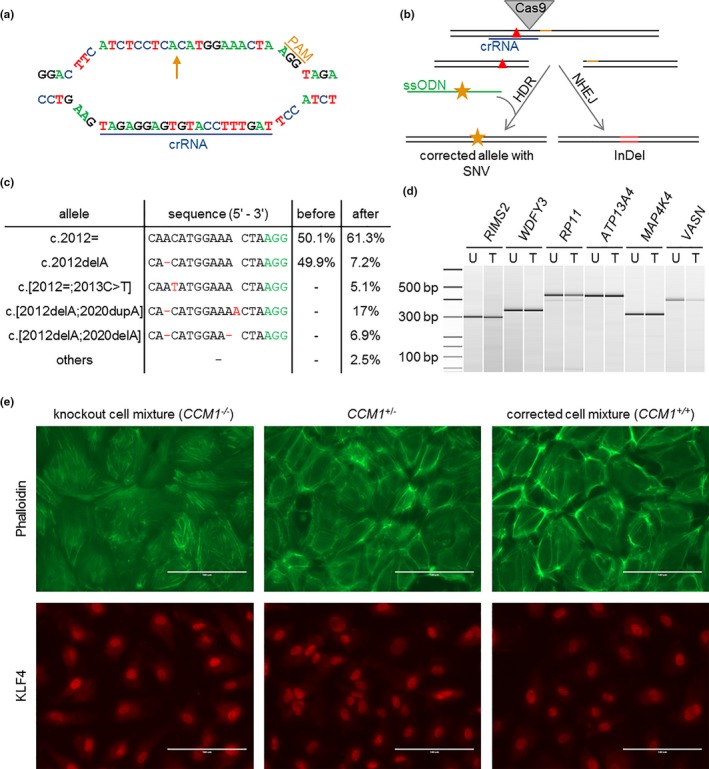
Correction of the patient‐specific pathogenic CCM1 variant and comparison of CCM1+/+, CCM1+/− and CCM1−/− BOECs by immunofluorescence. (a) crRNA and PAM sequence at the CCM1 allele c.2012delA (arrow). (b) Strategy of HDR‐mediated correction of a pathogenic CCM1 variant (red triangle) with CRISPR/Cas9 genome editing. ssODN = single‐strand oligodeoxynucleotide, orange star = silent SNV. (c) Amplicon deep sequencing results after CRISPR/Cas9 genome editing. The variant read frequencies before and after CRISPR/Cas9 transfection are given next to the sequence alignments. (d) T7 endonuclease I cleavage of PCR products from untreated (U) and treated (T) cells spanning possible off‐target sites. Ladder bands are given in base pairs (bp). (e) Characterization of the knockout cell mixture (left), the unmodified, patient‐derived BOECs (middle), and the corrected cell mixture (right). Visualization of the cytoskeleton using phalloidin reveals a regular F‐actin organization without the formation of stress fibers in the patients‐derived and corrected BOEC mixtures while CCM1‐/‐ cells demonstrated stress fiber bundles (upper panel). The strongest expression of the transcription factor KLF4 can be found in the knockout cell mixture when compared to unmodified and corrected BOECs (lower panel)
