Figure 5.
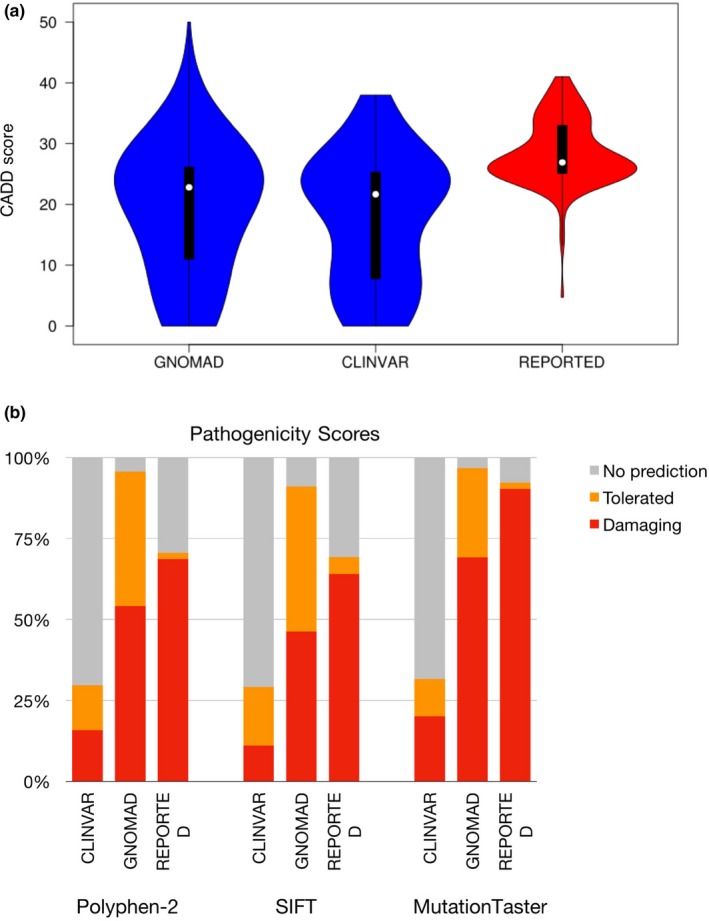
(a) Violin plot comparing the CADD scores of variants in the glycophosphatidylinositol (GPI) biosynthesis genes for Clinvar's benign variants, gnomAD's missense singletons and 145 reported variants. T‐tests suggest statistical difference between the reported variants and the Clinvar variants (ρ < 2.2e‐16) as well as with gnomAD's singletons (ρ = 1.99e‐06). (b) Comparison of the pathogenicity predictions by Polyphen‐2, SIFT, and MutationTaster for Clinvar's benign variants, gnomAD's missense singletons and the GPI biosynthesis genes‐reported variants. Any prediction considered as damaging or potentially damaging was included in the damaging category. The tolerated category includes anything predicted as tolerated, benign, probably benign. The NO prediction category consists of the variants for which the tool could not compute a prediction
