SUMMARY
The Saccharomyces cerevisiae (Sc) R2TP complex affords an Hsp90-mediated and nucleotide-driven chaperone activity to proteins of small ribonucleo-protein particles (snoRNPs). The current lack of structural information on the ScR2TP complex, however, prevents a mechanistic understanding of this biological process. We characterized the structure of the ScR2TP complex made up of two AAA+ ATPases, Rvb1/2p, and two Hsp90 binding proteins, Tah1p and Pih1p, and its interaction with the snoRNP protein Nop58p by a combination of analytical ultracentrifugation, isothermal titration calorimetry, chemical crosslinking, hydrogen-deuterium exchange, and cryoelectron microscopy methods. We find that Pih1p-Tah1p interacts with Rvb1/2p cooperatively through the nucleotide-sensitive domain of Rvb1/2p. Nop58p further binds Pih1p-Tahp1 on top of the dome-shaped R2TP. Consequently, nucleotide binding releases Pih1p-Tah1p from Rvb1/2p, which offers a mechanism for nucleotide-driven binding and release of snoRNP intermediates.
In Brief
Tian et al. report that AAA+ ATPases, Rvb1/2p, though previously shown to form both dodecamer and hexamer, form a hexameric base for binding co-chaperone Pih1p-Tah1p. Tian et al. also showed that the binding interface is made of flexible and nucleotide-sensitive insertions within Rvb1/2p, which offers the basis for nucleotide-driven client release.
Graphical abstract
INTRODUCTION
The R2TP complex is a four-protein complex required for assembly and localization of small nucleolar ribonucleoprotein (snoRNP) complexes (Boulon et al., 2008; Kakihara and Houry, 2012; Zhao et al., 2005, 2008). In yeast, the R2TP complex is composed of four proteins: Rvb1p (molecular weight [MW] = 50.5 kDa), Rvb2p (MW = 51.6 kDa), Pih1p (YHR034C, MW = 39.5 kDa), and Tah1p (YCR060W, MW = 12.5 kDa). Rvb1p and Rvb2p are two essential AAA+ (ATPase-associated diverse cellular activities) proteins that share features with the holiday junction resolving protein RuvB (Putnam et al., 2001). Rvb1/2p were first implicated in snoRNP maturation after they were co-purified with a poly(A)-tagged U14 snoRNP from mouse nuclear extract (Newman et al., 2000), and subsequently shown to be required in yeast for snoRNP stability and nucleolus localization (King et al., 2001). Remarkably, Rvb1/2p were also independently identified to be part of the chromatin remodeling complexes, INO80 (Shen et al., 2000) and SWR-C (Krogan et al., 2003), at about the same time and were later shown to form the ring-shaped head of the two large and flexible complexes (Nguyen et al., 2013; Tosi et al., 2013; Watanabe et al., 2015). Label-free quantitative proteomic analysis of complexes associated with TAP-tagged yeast Rvb1/2p revealed that R2TP is the third most abundant Rvb1/2p-associated complex next to INO80 and SWR-C (Lakshminarasimhan et al., 2016). Differing in both composition and function from the chromatin remodeling complexes, however, R2TP functions with the molecular chaperone heat-shock protein 90 (Hsp90) in assisting snoRNP assembly and localization (Boulon et al., 2008; Zhao et al., 2008). In fact, Pih1p (protein interaction with Hsp90) and Tah1p (TPR-containing protein associated with Hsp90) were initially identified through yeast two-hybrid screening with Hsp90 or the box C/D snoRNP protein Nop58p (Gonzales et al., 2005; Zhao et al., 2005) and Tah1p was later confirmed to interact specifically with Hsp90 (Back et al., 2013; Zhao et al., 2005, 2008). Although Pih1p or Tah1p is not essential to cell growth (Zhao et al., 2005), double disruption of Pih1p with another snoRNP biogenesis factor, Rsa1p or Hit1p, exhibits synthetic lethality (Rothe et al., 2014). Depletion or deletion of three of the four R2TP subunits in yeast cells reduces the level of small nucleolar RNAs (snoRNAs) and impairs localization of snoRNP proteins (Gonzales et al., 2005; King et al., 2001; Rothe et al., 2014; Zhao et al., 2008). Consequently, mature 25S and 18S rRNA and ribosome decrease in abundance while 35S and 23S intermediate accumulate in these cells (Gonzales et al., 2005; King et al., 2001; Rothe et al., 2014; Zhao et al., 2008).
How R2TP/Hsp90 facilitates snoRNP maturation remains unclear. The current model suggests that Pih1p-Tah1p is an adaptor to Rvb1/2p for snoRNP intermediates. Multiple evidence from both in vivo and in vitro studies supports a specific interaction between Pih1p and unassembled Nop58p (Gonzales et al., 2005; Kakihara et al., 2014; McKeegan et al., 2009; Prieto et al., 2015; Quinternet et al., 2015). Nop58p forms, with its close homolog Nop56p, the scaffold of all box C/D snoRNPs onto which individual Snu13p-bound box C/D RNAs dock (Liang and Li, 2011). The Nop58p-containing U3 snoRNP is a core of the 90S pre-ribosome responsible for rRNA processing, whose assembly was recently visualized (Chaker-Margot et al., 2017; Kornprobst et al., 2016; Sun et al., 2017). Binding of Nop58p to R2TP could prevent Nop58p from premature assembly in nucleoplasm. Consistently, in Pih1p-depleted cells, U3 snoRNA is observed to have a stronger association with Nop58p (Gonzales et al., 2005; Prieto et al., 2015), which explains impaired rRNA processing in these cells. In humans, the R2TP/Hsp90 system is also responsible for maturation of RNA polymerase II (Boulon et al., 2010; Forget et al., 2010), pseudouridine synthase NAP57/dyskerin (Machado-Pinilla et al., 2012), PIKKs complexes (Horejsi et al., 2010; Izumi et al., 2010; Takai et al., 2010), and the spliceosome (Bizarro et al., 2015). Delineation of the molecular interactions underlying R2TP assembly and its binding to clients is thus required to fully explain the function of R2TP.
Previous structural studies of isolated Rvb1/2p, Pih1p and Tah1p, and their interaction with Hsp90 have provided insights on features of the individual components (Figure 1) but did not result in any model of the assembled R2TP. Rvb1/2p share the core structure of AAA+ proteins and also contain Rvb-specific features (Huen et al., 2010; Jha and Dutta, 2009; Tucker and Sallai, 2007). The core AAA+ module is made up of domains I (DI) and III (DIII) that includes the conserved ATPase features (Walker A and B motifs, sensor domains 1 and 2, and the arginine finger) (Figures 1 and S1). The Rvb1/2p-specific domain, or domain II (DII), is a ~100-amino-acid insertion that includes the nucleotide binding (OB) fold (DII external, or DIIEXT) and a largely disordered loop (DII internal, DIIINT) (Figure 1A). Mutations in their Walker A and B motifs cause deleterious effects in multiple pathways or cell growth (Nano and Houry, 2013). Significantly, a 3-amino-acid in-frame insertion in DII of zebrafish Rvb2 (zfRvb2) led to development of hyperplastic embryonic hearts in the organism and, owing to the high degree of sequence homology, similar disruption in human RUVB2 DII is predicted to be lethal (Rottbauer et al., 2002). Pih1p is characterized by three distinct regions: an N-terminal PIH domain (34–166, Pih1p_NTD), an insertion with two intrinsically disordered regions (IDRs), and a C-terminal domain (268–344, Pih1p_CTD) (Figure 1B). Tah1p is composed of an N-terminal domain (1–91) containing two TPR (the Tetra-tricopeptide repeats) motifs and a C-terminal segment (92–111) with which Pih1p_CTD interacts (Pal et al., 2014; Quinternet et al., 2015). Hsp90 also interacts with Tah1p through its C-terminal MEEVD motif, specifically with both of TPR motifs of Tah1p (Back et al., 2013; Pal et al., 2014). As a result, Hsp90, Pih1p, and Tah1p can form a stable ternary complex with a 2:2:2 stoichiometry independent of R2TP assembly (Pal et al., 2014), and this complex stabilizes Pih1p (Zhao et al., 2008). Although the domain structures and most of the pairwise interactions are expected to remain, changes may take place as R2TP assembles into its functional form.
Figure 1. Schematic Representation of ScR2TP Protein Features.
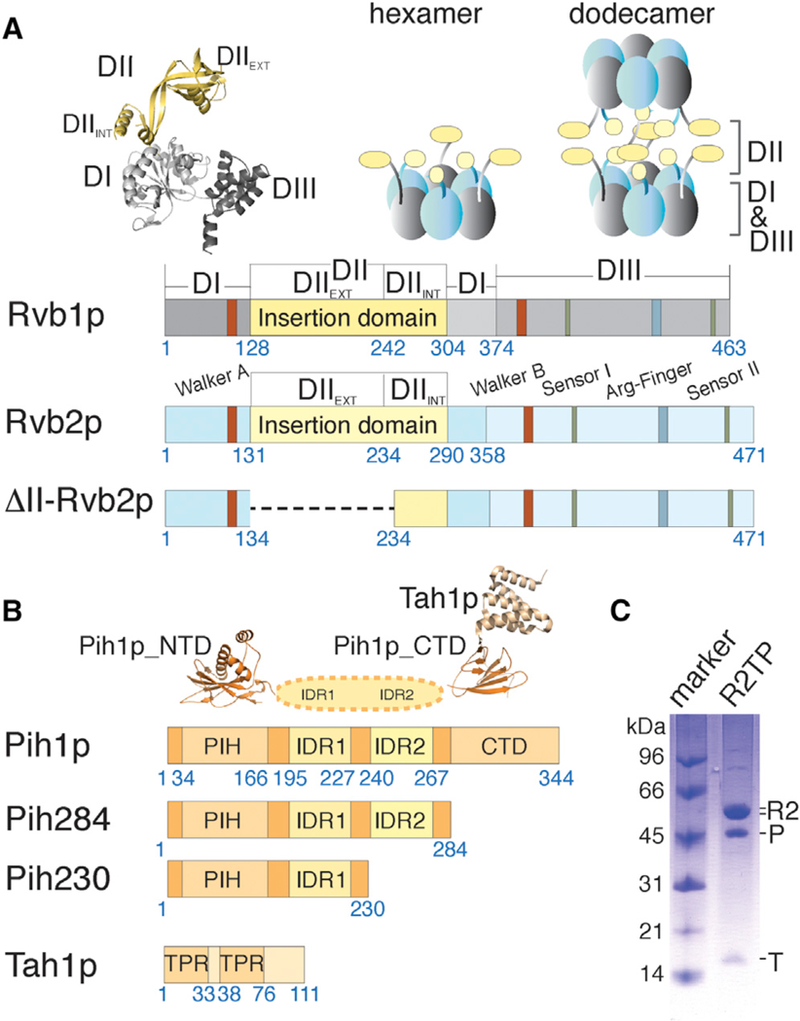
(A) Schematic diagrams depicting primary structural features of yeast Rvb1p and Rvb2p including both wild-type and mutant constructs used in this study. Residue numbers at the beginning and ends of each region discussed in the text are indicated under the diagrams. The crystal structure of human RUVBL1 (PDB: 1C9O) shown in ribbons illustrates the three conserved domains for both Rvb proteins colored as in the diagrams. Schematic arrangements of previously observed oligomers, heterohexamer and heterododecamer, for the yeast Rvb1/2p complex are illustrated above the diagrams by solid blocks in gray and light blue.
(B) Schematic diagrams depicting primary structural features of yeast Pih1p and Tah1p. Residue numbers at the beginning and ends of each region discussed in the text are indicated under the bar diagrams. The available crystal structures for the N-terminal domain of Pih1p (Pih1p_NTD) (PDB: 4CHH) and Tah1 p bound to the C-terminal domain of Pih1p(Pih1p_CTD) (PDB: 4CGU) are shown above the diagrams.
(C) Co-purified Saccharomyces cerevisiae ScR2TP complex from recombinantly expressed components is shown on an SDS-PAGE gel with a molecular weight marker. “R2” denotes the co-purified, tag-free Rvb1/2p complex (see a zoomed view in Figure S2), “P” denotes His-tagged Pih1p, and “T” denotes tag-free Tah1p. The stoichiometry of the co-purified ScR2TP was determined by label-free mass spectrometry to be ~0.5:1.6:3:3 (Table S1).
Structural studies of Rvb1/2p-associated complexes are confounded by weak and non-specific interactions (Nano and Houry, 2013), the presence of IDRs (Paci et al., 2012), and the multiple oligomeric forms of Rvb1/2p (Cheung et al., 2010a). A combination of techniques with different strength is required to gain the full range of possible structures. In previous Rvb1/2p structural studies, both X-ray crystallographic and cryoelectron microscopy (cryo-EM) methods were employed that revealed two possible oligomeric states. Crystal structures of the human RVB1/RVB2 proteins (67% sequence identity to yeast) and a Chaetomium thermophilum (Ct) Rvb1-Rvb2 complex (69% sequence identity to yeast) showed either a single or a double ring structure: a heterohexamer containing alternating RVB1 and RVB2 or a dodecamer with two stacked hexameric rings (Lakomeket al., 2015; Matias et al., 2006; Niewiarowski et al., 2010) (Figure 1A). The ATPase core forms the base of each hexamer ring and DII protrudes from the base to form a secondary ring on top (Lakomek et al., 2015; Matias et al., 2006; Niewiarowski et al., 2010) (Figure 1A). Electron microscopy (EM) studies of either negatively stained or ice-embedded Rvb1/2 yielded only dodecamers of Rvb1/2 owing to the single preferred orientation of the hexamer on carbon-coated sample grids (Cheung et al., 2010b; Gribun et al., 2008). How Pih1p-Tah1p interacts with Rvb1/2p and with which oligomeric form has been recalcitrant to studies by these structural methods.
The important question of how Rvb1/2 utilizes its nucleotide binding and ATPase activity in snoRNP chaperone function remains largely unanswered. Structural studies of isolated Rvb1/2 clearly show that DII of both Rvb1 and Rvb2 undergo a rotational change upon nucleotide binding that is further enhanced by ATP hydrolysis (Ewens et al., 2016; Jeganathan et al., 2015; Lakomek et al., 2015). This conformational change causes a relative rotation of the two stacks of the dodecamer, as DII mediates the contacts between the stacks (Ewens et al., 2016). The implication of the nucleotide-driven conformational change in R2TP function, however, remains unknown.
Here, we describe structural characterization of recombinantly expressed and assembled yeast ScR2TP complex (Figure 1C) and its interaction with snoRNP protein Nop58p, based on analytical ultracentrifugation (AUC), chemical crosslinking, hydrogen-deuterium exchange (HDX), EM single-particle reconstruction, and mutational studies. We overcome sample preparation and preferred orientation problems in cryo-EM studies and show structural model of fully assembled ScR2TP. We identify the binding interfaces within and on the ScR2TP complex and suggest a structural mechanism underlying the nucleotide-driven functional cycle.
RESULTS
Pih1p-Tah1p Drives Formation of Rvb1/2p Heterohexamer
To firmly establish the oligomerization state of Rvb1/2p within ScR2TP, we first performed sedimentation velocity experiments with individually purified ScR2TP components (Figure S2) in an AUC analysis. Under the condition we used, the isolated Rvb1/2p complex exhibits a nearly equal distribution of two major sedimentation species centered around sedimentation coefficients 10.5 S and 18.5 S (Figure 2A). These values are consistent with those of Rvb1/2p hexamer and dodecamer estimated from the bead model corresponding to crystal structures of human Rvb1 hexamer (10.3 S) and Ct Rvb1/2 dodecamer (17.1 S) (Garcia De La Torre et al., 2000). Significantly, in the presence of molar excess Pih1p-Tah1p, the dodecamer species of Rvb1/2p (18.5 S) was undetectable whereas the hexamer species (10.5 S) shifted to 12.4 S with increased abundance. The emergence of a new 3.1 S species likely corresponds to the unbound Pih1p-Tah1p heterodimer (Figure 2A). The 12.4 S species indicates formation of the ScR2TP complex from the Rvb1/2p hexamer at the expense of Rvb1/2p dodecamer dissociation (Figure 2A).
Figure 2. Biophysical Characterization of ScR2TP Complex.
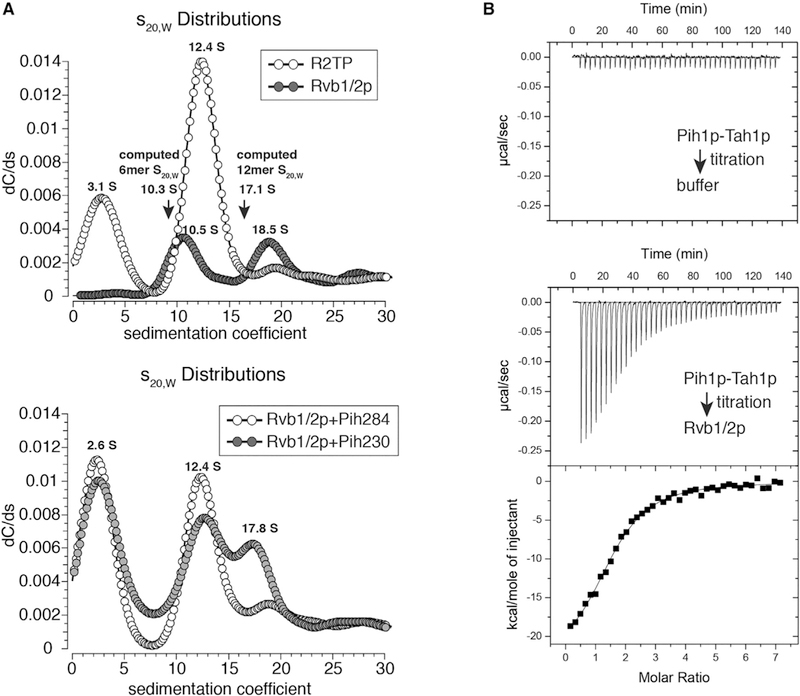
(A) Monte Carlo predictions combined with decreasing sigmoidal spectrum analysis of sedimentation velocity data for ScR2TP (A) and Pih1p fragments incubated with Rvb1/2p (B) compared with that of isolated Rvb1/2p. The computed sedimentation coefficients (arrows) were obtained from Zeno by use of US-SOMO modeled Rvb1/2p structure (see STAR Methods). Upper: comparison of sedimentation coefficient (S20, water) distribution forScR2TP (open circles) and isolated Rvb1/2p (solid circles) indicates a clear shift from the18.5Sspecies (dodecamer) tothe10.5Sspecies (hexamer) upon ScR2TPformation. Lower: Comparison of sedimentation coefficient (S20, water) distribution for Rvb1/2p incubated with Pih284 (open circles) and Rvb1/2p incubated with Pih230 (solid circles) indicates that Pih284 is sufficient to shift the dodecamer (17.8 S) to the hexamer (12.4 S) species.
(B) Example isothermal titration calorimetry profiles and data analysis of Pih1p-Tah1p complex titrating into buffer or an Rvb1/2p solution, respectively. The binding isotherm resulted from integrating successive heat release peaks was fitted by use of several binding models and that of two serial binding sites was chosen. Molar ratios shown were those ofPih1p-Tah1p hetero dimer versus Rvb1/2p hexamer. Thermodynamic parameters were computed from the averages of four separate titration trials. KD values are in nM. ΔH and –TΔS are in kcal/mol.
To map the region of Pih1p-Tah1p complex required for the oligomeric shift of Rvb1/2p, we performed additional sedimentation velocity experiments with Rvb1/2p in complex with two Pih1p fragments (Figure S2). The 1–230 fragment alone was insufficient to fully induce the dodecamer to hexamer shift whereas the 1–284 fragment did (Figure 2A), suggesting that the second IDR (IDR II, 240–267) of Pih1p is important for this function. The role of IDR II in binding Rvb1/2p was also observed previously in a pull-down study (Paci et al., 2012).
Isothermal Titration Calorimetry Measurement Reveals Cooperative Assembly
We characterized the thermodynamics and stoichiometry of Pih1p-Tah1p association with Rvb1/2p hexamer by isothermal titration calorimetry (ITC). The simplest model that gives rise to a good fit to the binding isotherm assumes a double serial binding event (Figure 2B). The first Pih1p-Tah1p binding is strong with an apparent dissociation constant of KD1 = 155 nM, which is dominated by large enthalpy change. The second binding is much weaker with an apparent dissociation constant of KD2 = 1.2 μM, indicating a negative cooperativity (Figure 2B). The second binding event is also dominated by a large enthalpy change, suggesting a contribution of hydrogen bonding and van der Waals interactions to Pih1p-Tah1p association with Rvb1/2p.
The serial two-site binding model is in agreement with our stoichiometry measurement by label-free mass spectrometry (Table S1). Mass spectrometry analysis of isolated ScR2TP complex reveals an average stoichiometry of 0.5:1.6:3:3 for Tah1p:Pih1p:Rvb1p:Rvb2p. The association of between one and two Pih1p with the Rvb1/2p hexamer reflects one tight and one weak binding site detected by ITC. It is not known whether the substoichiometric amount of Tah1p detected by mass spectrometry is physiological or a result of our purification process.
Structural Overview of the ScR2TP Complex
We next purified the ScR2TP complex by an affinity method (Figure S2) and studied its structure by both negative stain and cryo-EM methods. At a low concentration suitable for negative stain analysis, the ScR2TP complex showed particles largely characteristic of hexamers (Figure 3), but unlike previously observed Rvb1/2p hexamers with a single preferred top-view orientation (Cheung et al., 2010b; Gribun et al., 2008). Two-dimensional (2D) classification displayed averages with both disc-like end-on and rice bowl-shaped side views, similar to those observed for SWR-C (Watanabe et al., 2015), which enabled us to perform single-particle reconstruction. Three-dimensional (3D) classification and refinement revealed that the particles were heterogeneous with varying amount of electron density on top of the disc (Figure S3). Iterative sorting based on both 2D and 3D classifications led finally to two 3D classes that we interpret as ligand-bound and-unbound (or weakly bound) classes (Figure S3).
Figure 3. Structural Analysis of ScR2TP.
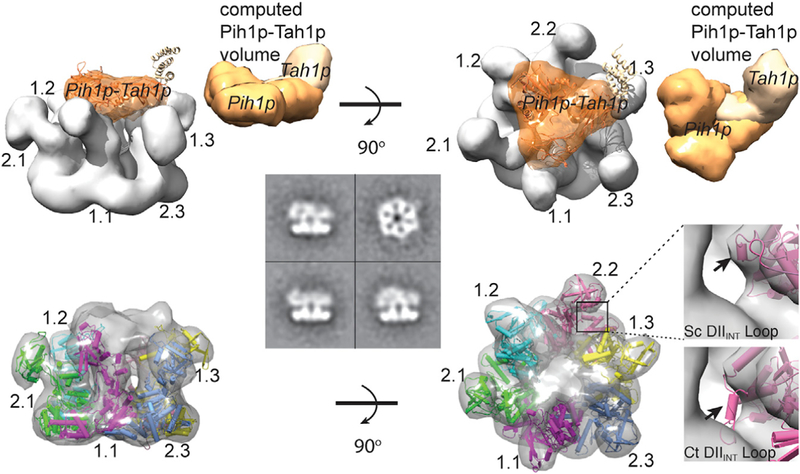
Cryo-EM maps of the ScR2TP complex shown as surface models at two orthogonal views. Water-shed segmentation electron density is shown above and the electron density fitted with refined Rvb1/2p structure is shown below. Four representative 2D class averages for negatively stained particles are also shown at the center, including a top view and three side views with varying central mass. The segment identified as bound Pih1p-Tah1p is colored orange. A model of the full-length Pih1p-Tah1p complex was generated from the crystal structures of the Pih1p N-terminal domain (PDB: 4CHH) and the Pih1p C-terminal domain bound with Tah1p (PDB: 1CGU), and a modeled random coil structure of the intrinsically disordered regions of Pih1p. For the purpose of structural comparison, the Pih1p-Tah1p model is placed into the segmented cryo-EM density of ScR2TP with an arbitrary orientation and its computed volume displayed on the side. Each Rvb1/2p chain is represented by a unique color indicated by 1.1–1.3 for Rvb1p.1-Rvb1p.3and 2.1–2.3for Rvb2p.1-Rvb2p.3. The inset shows an example (DIIINT) of the observed difference between the isolated Chaetomium thermophilum (Ct) Rvb1/2p and the Saccharomyces cerevisiae Rvb1/2p in the ScR2TP complex. Arrows indicate the direction of rotations of the models.
We then collected a cryo-EM dataset of ice-embedded ScR2TP prepared at a higher concentration than that used for negative stain analysis. A small fraction of dodecamers was observed, whereas most particles remained hexameric (Figure S3). The dodecamers were eliminated by 2D classification leaving 54,712 particles for 3D reconstruction. These were subjected to rounds of 3D classification with several symmetry considerations (Figure S3). The C3 symmetry was found to be necessary at early refinement stages, resulting in five classes of varying degrees of heterogeneity. The class displaying the best-resolved features was selected for further refinement with pseudo-C3 symmetry as implemented in the program Frealign (Grigorieff, 2016). This procedure resulted in three classes with subtle differences in the quality of the DII and the central electron density. The one that had the best-resolved features overall was selected as the final reconstruction from 4,839 particles with a final resolution of 14 Å (Fourier shell correlation 0.143) (Figure S3). This structure was segmented by use of the watershed segmentation feature available in UCSF Chimera (Pettersen et al., 2004), to produce segmented electron densities corresponding to a base formed by the Rvb1/2p heterohexamer and a cap associated with the bound Pih1p-Tah1p (Figure 3).
We constructed several atomic model structures of the yeast Rvb1/2p heterohexamer by homology building based on the crystal structure of Ct Rvb1/2 heterohexamer (PDB: 4WVY) or human Rvb1 (PDB: 2C9O). We placed the resulting models of ScRvb1/2p hexamer into the refined EM density, keeping the same relative orientations of DII for Ct Rvb1 and Ct Rvb2, and subjected them to the density-guided refinement procedure implemented in Rosetta (DiMaio et al., 2015). The process was iterated multiple times with different starting models. The final refined model was found to be independent of the template and fitted well with the EM density (Figure 3).
The density over the placed Rvb1/2p can be readily assigned to the bound Pih1p-Tah1p, although an atomic model of the heterodimer is not possible due to limited resolution. However, features of the bound Pih1p-Tah1p can be gleaned from the electron density. Clearly, Pih1p-Tahp1 is located on top of the Rvb1/2p heterohexamer and directly contacts DII of the Rvb1/2p proteins (Figure 3). The density can accommodate one Pih1p-Tahp1 complex based on its estimated volume from known crystal structures of the fragments (Figure 3), although possible “contamination” to the density due to some misaligned particles makes this estimation inaccurate. Notably, the model is in agreement with the stoichiometry of Pih1p-Tah1p association with Rvb1/2p measured by ITC and mass spectrometry.
Structural Evidence for ScR2TP Dissociation upon ATP/ ADP Binding
By taking advantage of the distinctly different orientations in EM of raw particles between ScR2TP and free Rvb1/2p, we visualized the impact of nucleotide binding and hydrolysis on ScR2TP structure. We incubated the ScR2TP sample used for cryo-EM analysis with molar excess ATP and ADP, respectively, at 30°C for 30 min and imaged the treated ScR2TP complexes by negative-stain EM. 2D classification of ~5,000 particles in both cases yielded 2D averages mostly in top views (Figure S4) that are characteristic of free Rvb1/2p hexamers (Cheung et al., 2010b; Gribun et al., 2008) but not assembled ScR2TP. This result is consistent with the fact that Rvb1/2p did not co-purify with bead-immobilized Pih1p in the presence of ADP or ATP but could in their absence (Kakihara et al., 2014), thereby supporting a nucleotide-driven dissociation mechanism.
ScR2TP Assembly Depends on the Insertion Domain and Oligomerization of Rvb1/2p
To further confirm the importance of DII of Rvb1/2p in binding Pih1p-Tah1p in solution, we separately purified the wild-type Rvb1/2p complex, Rvb1p, Rvb2p, and a DII-truncated Rvb2p (deleting residues 134–234) (truncation of DII of Rvb1p led to insoluble protein) and studied their individual assembly with the Pih1p-Tah1p complex (Figure 4). We incubated non-tagged Rvb proteins with His-tagged Pih1p-Tah1p and loaded the mixture onto a nickel-Ni-nitrilotriacetic acid column (Figure 4A). We found that Pih1p-Tah1pco-purified efficiently with a DII-truncated (in Rvb2p) Rvb1/2p (R2TP-ΔII2), as well as Rvb2p alone (R22TP) (Figures 4A and 4B). However, it did not co-purify with DII-truncated Rvb2p mutant (R22TP-ΔII) (Figure 4B), suggesting an importance of DII and that its presence on either Rvb1p or Rvb2p is sufficient for ScR2TP assembly. To further test whether isolated DII is capable of binding Pih1p-Tah1p, we constructed and purified DII of Rvb2p (DII2) for co-purification studies. Even in large excess, DII2 did not bind Pih1p-Tah1p (Figure 4A), suggesting that Pih1p-Tah1p binding also requires assembled Rvbs. Consistently, Pih1p-Tah1p did not co-purify with Rvb1p (Figure 4C), which was shown to form either aggregates or monomer at the concentration used for negative-stain EM (Figure S2C).
Figure 4. Biochemical Verification of the Importance of Insertion Domain in ScR2TP Assembly.
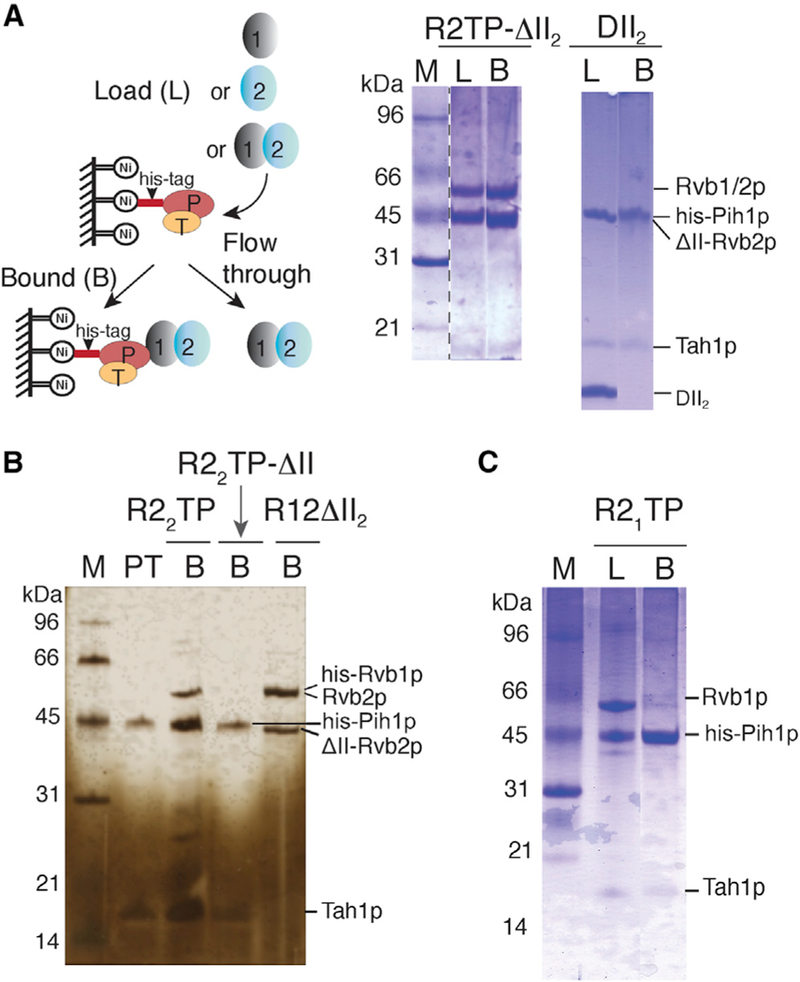
Solid ovals represent Rvb1p (1), Rvb2p, or Rvb2p DII deletion (2), Pih1p (P), and Tah1p (T). SDS-PAGE gels are stained by either Coomassie brilliant blue (blue) or silver nitrate (yellow). Samples analyzed on gels are denoted as L for load, B for bound, PT for Pih1p-Tah1p control, and M for protein markers.
(A) Schematic representation of the assembly and pull-down method (left) and SDS-PAGE gel analysis of Pih1p-Tah1p binding to Rvb1/2p lacking domain II in Rvb2p (ScR2TP-ΔII2) or DII2 (domain II of Rvb2p) (right).
(B) SDS-PAGE gel (silver-stained), showing Pih1p-Tah1p binding to Rvb2p (R22TP) but not to Rvb2p lacking its domain II (R22TP-ΔII). The last lane (R12-ΔII2) shows binding of Rvb1p to the same Rvb2p lacking its domain II (Rvb2p-ΔII) sample used for the R22TP-ΔII complex formation.
(C) SDS-PAGE gel, showing formation of the Pih1p-Tah1p-Rvb1p complex (R1TP).
We then asked whether R22TP complex is hexameric by carrying out negative-stain EM analysis. The high yield of the R22TP complex (Figure S2) allowed us to apply the Grafix procedure for further separation of different species on a glycerol gradient (Stark, 2010) before making grids. Single-particle reconstruction resulted in two major classes with varying amounts of central densities (Figures 5 and S5). To take advantage of the three-fold symmetry in Rvb1/2p, we obtained two 3D classes, one with pseudo-C3 (Figure 5B) and one without symmetry (Figure 5C) constraint refinement. For each model, the hexameric ring could be unambiguously assigned to Rvb2p and the central density to the bound Pih1p-Tah1p. Unlike ScR2TP in which Pih1p-Tah1p makes extensive contacts with both DIIEXT and DIIINT of Rvb1p, Pih1p-Tah1p in R22TP contacts largely DIIINT of all six Rvb2p subunits. The fuller density on top observed in GraFix-separated and crosslinked R22TP (Figure 5C) is more comparable with the estimated volume of Pih1p-Tah1p (Figure 3) and likely a result of stabilized Tah1p.
Figure 5. Negatively Stained EM Structures for the ScR22TP Complex.
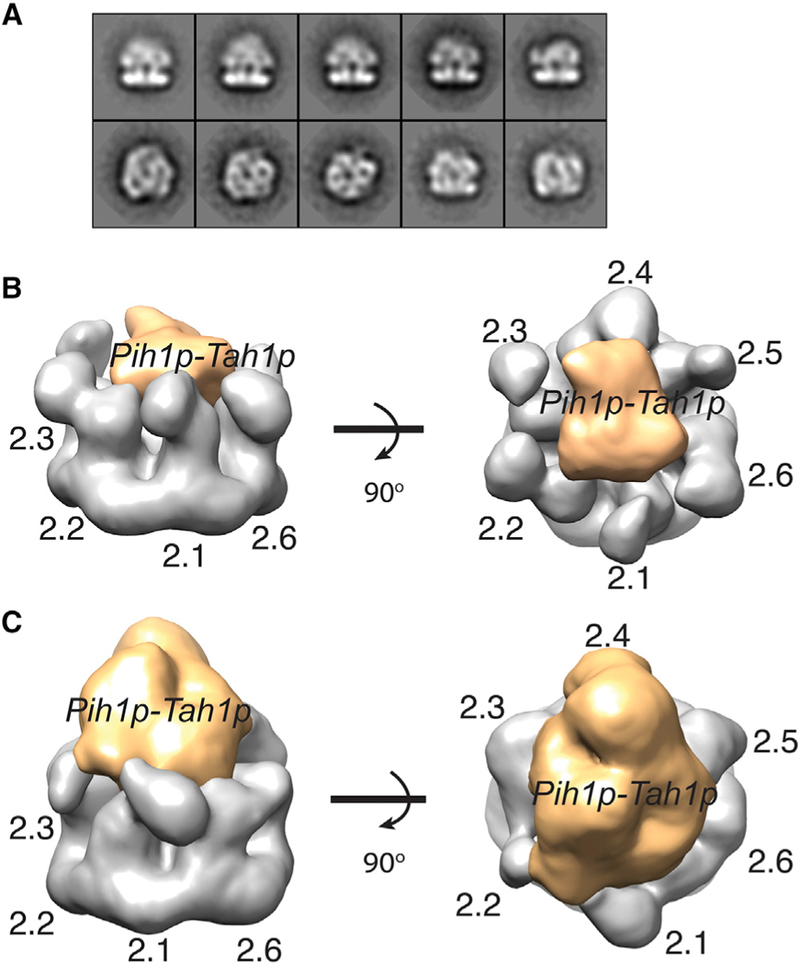
(A) Representative 2D class averages of the R22TP particles Indicating both top and side views.
(B) Watershed segmented electron density of the smaller 3D class shown at two orthogonal views. Orange: assigned to Pih1p-Tah1p. Gray: assigned to Rvb2p. Rvb2p are labeled as 2.1–2.6.
(c) Watershed segmented electron density of the bigger 3D class shown at two orthogonal views. Orange: assigned to Pih1p-Tah1p. Gray: assigned to Rvb2p. Rvb2p are labeled as 2.1–2.6.
Rvb1/2p Undergo Large Conformational Changes upon Pih1p-Tahp1 Binding
Superimposition of the ScRvb1/2p hexamer structure of the ScR2TP complex on that of the isolated CtRvb1/2 hexamer or human Rvb1 hexamer revealed pronounced differences among the structures in three regions. Two of these regions are located in the insertion domain of both Rvb1p and Rvb2p. The entire head domain, or DIIEXT, is repositioned in the ScR2TP complex structure relative to that in the Ct Rvb1/2 structure (Figures 3 and 6; Movie S1). Remodeling of DIIEXT is also accompanied by a large rotation of the DIIINT loop. In the Ct Rvb2 structure, DIIINT reaches into the insertion domain of its neighboring subunit, thereby enhancing the stability of the hexameric ring structure. In the modeled ScR2TP structure, however, DIIINT in each subunit is released from its ring-stabilization position through a ~90° rotation to engage the bound Pih1 p-Tah1 p on top of the Rvb ring (Figure 6 and Movie S1). The third region of difference is in the Rvb1/2p ring made up by their core domains DI and DIII. In the ScR2TP complex, the ring is significantly more open, with an ~8 Å increase in diameter from that of the Ct Rvb1/2 structure (Figure 6 and Movie S1).
Figure 6. Structural Remodeling in Rvb1/2p upon ScR2TP Formation.
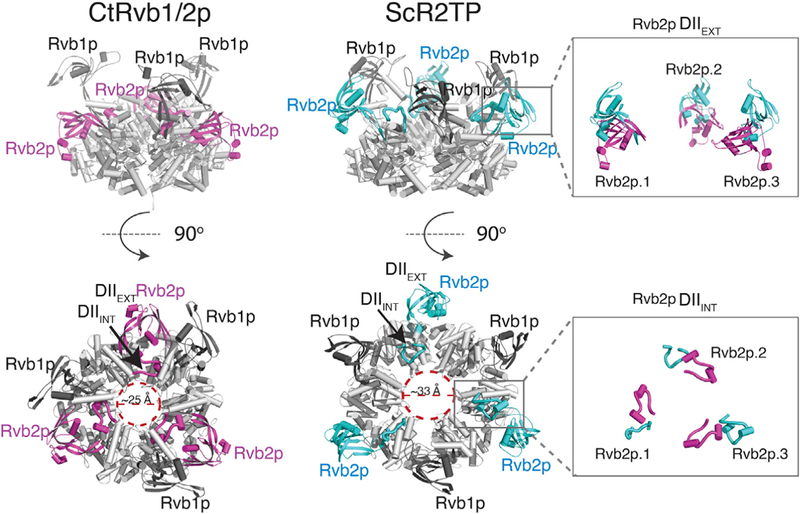
The Pih1p-Tah1p-bound Rvb1/2p heterohexamer structure resulted from electron density-guided refinement is compared side by side with the crystal structure of C. thermophilum (Ct) Rvb1/2p complex (PDB: 4WVY) in two orthogonal views. The AAA+ core for both complexes is colored in gray and the insertion domain of Rvb2p is colored magenta for Ct and teal for Sc. DIIINT and DIIEXT denote the insertion domain II internal and external elements (see definition in Figure 1) and are labeled for one Rvb2p subunit. Insets show comparisons of DIIEXT (upper) and DIIINT (lower) conformations between superimposed Ct (magenta) and the Pih1p-Tah1p-bound (teal) Rvb1/2p.
The observed remodeling in the three regions of the ScR2TP structure is consistent with our mutational data, which indicates that the insertion domain mediates Pih1p-Tah1p binding. The conformational remodeling also explains the role of Pih1p-Tah1p in shifting the Rvb1/2p oligomeric state, because DII was observed to be the interface in dodecamer formation (Ewens et al., 2016; Lakomek et al., 2015). Sequestration of DII by the bound Pih1p-Tah1p would prevent formation of the Rvb1/2p dodecamer.
The Intrinsically Disordered Region in Pih1p Contacts the Rvb1/2p Insertion Domain
To map the regions in Pih1p-Tah1p that contact Rvb1/2p in solution, we carried out lysine-specific chemical crosslinking followed by mass spectrometric identification of crosslinked peptides (Schmidt and Robinson, 2014). The ScR2TP sample used for cryo-EM studies was directly incubated with deuterated (d4) or non-deuterated (d0) bis(sulfosuccinimidyl)suberate (BSS) before being combined and loaded onto an SDS-PAGE gel. The crosslinked species were excised from the gel and treated with trypsin. The resulting peptide fragments were analyzed for intermolecular crosslinks whose masses were simultaneously modified by BSS-d0 and BSS-d4. The crosslinks between Rvb1p and Rvb2p fitted nicely to the modeled heterohexamer of Rvb1/2p (Table S2) and thus provided a positive control for examining Pih1p-Tah1p crosslinks. Rvb1/2p crosslinked exclusively to Pih1p at its N terminus (Lys10 and Tyr19) and IDR (Lys239, Lys248, Lys275) but not to any regions of Tah1p (Figures 7A and S6). Furthermore, the regions of Rvb1p that cross-linked to Pih1p were mapped exclusively to its insertion domain (Lys171, Lys174, and Lys177). Note that three lysine residues of Rvb2p insertion domain, Lys157, Lys198, and Lys183, also crosslinked to Pih1p IDR (Lys275) and the N terminus (Lys10) (Figures 7A and S6). This result reflects the possibility of some Rvb2p-only hexamers in solution or an expanded binding interface at a small number of ScR2TP particles not represented in our 3D reconstruction. This possibility is consistent with our observation that Pih1p-Tah1p readily associates with Rvb2p but not Rvb1p (Figure 4).
Figure 7. Biochemically Determined Interfaces and Architecture of Nop58-ScR2TP Complex.
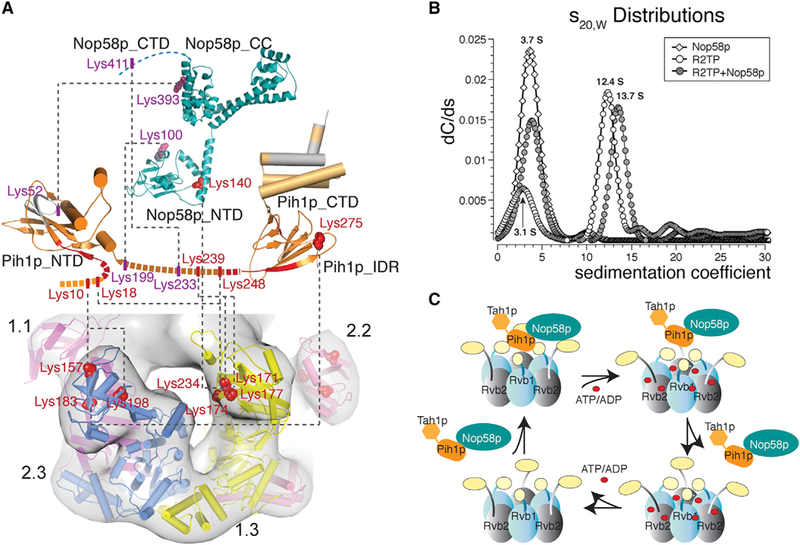
(A) Mapped interface between the Pih1p-Tah1p complex and the Rvb1/2p complex and between Nop58p_447 and ScR2TP obtained from lysine-specific chemical crosslinking and hydrogen/deuterium exchange methods. Proteins are represented by cartoons colored by subunits and lysine residues forming inter-subunit crosslinks are shown as either red spheres or sticks. Cryo-EM map is shown together with the refined Rvb1/2p hexamer structure. Black dashed lines indicate the cross linked pairs. Intra-molecular cross links or those between Rvb1pand Rvb2pand between Pih1p-Tah1pare not shown but are listed in TableS2. Hydrogen/deuterium exchange results were obtained for Pih1p-Tah1p in the presence and absence of Rvb1/2p complex. Residues with decreased deuterium uptake are colored red and those with increased deuterium uptake are colored blue.
(B) Monte Carlo predictions combined with decreasing sigmoidal spectrum analysis of sedimentation velocity data for Nop58p_447-ScR2TP complex. Comparison of sedimentation coefficient (S20, water) distribution for ScR2TP (open circles) and Nop58p_447-ScR2TP complex(solid circles) indicates a clear shift from the 12.4 S species to the 13.7 S species upon Nop58p binding.
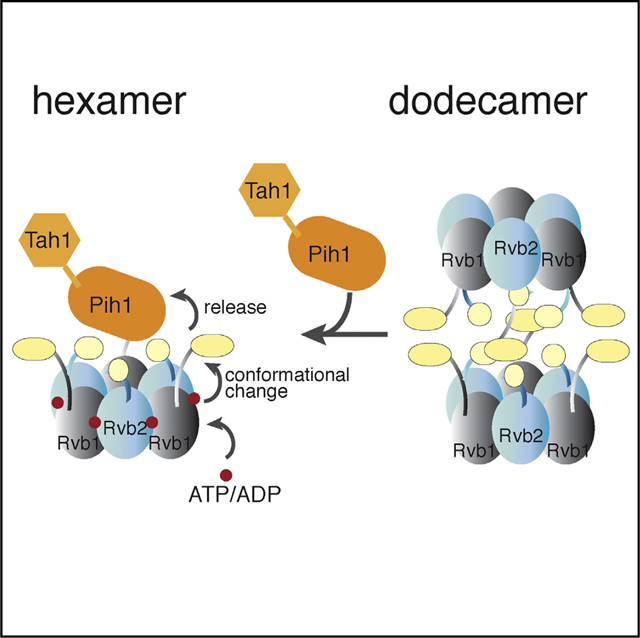
(C) A cartoon model of the ScR2TP and ScR2TP-Nop58p complex structures and the structural basis for Nop58p release in a nucleotide-dependent cycle. Binding of nucleotides (ATP/ADP) causes a rotation in the insertion domain of Rvb1/2p, which leads to distortion of Pih1p-Tah1p binding site and therefore release of Pih1p-Tah1p along with Nop58p. Release of nucleotides returns Rvb1/2p to the state for Pih1p-Tah1p (and Nop58p) binding again.
Independent HDX experiments yielded consistent interaction results. Pih1p residues 25–35 (N terminus) and 261–270 (IDR2) exhibited a clear decrease in solvent accessibility when mixed with Rvb1/2p, again indicating that these regions are involved in binding Rvb1/2p consistent with their direct crosslinks to Rvb1/2p (Figure 7A). Similarly, no residues of Tah1p showed decreased solvent accessibility upon Rvb1/2p binding. Significantly, HDX also detected an increase in solvent accessibility in residues 38–60 of Pih1p and 1–44 of Tahp1p (Figures 7A and S7), suggesting a rearrangement of these regions upon formation of the ScR2TP complex.
Taken together, cryo-EM, mutagenesis, chemical crosslinking, and HDX data showed remarkable agreement and established that the Rvb1/2p insertion domain contacts the N terminus and the intrinsically disordered region of Pih1p in the ScR2TP complex. Based on electron density volume comparison and the measured stoichiometry, we propose a model of an assembled ScR2TP complex in which one Pih1p-Tah1p complex is bound atop the Rvb1/2p heterohexamer mediated by Pih1p-Rvb1/2p contacts.
ScR2TP Interaction with the Client Protein Nop58p
We next addressed how ScR2TP interacts with its client protein, Nop58p of box C/D snoRNPs, by carrying out lysine-specific crosslinking and analytic ultracentrifugation. We used a truncated Nop58p (residues 1–447), Nop58p_447, in which the dispensable and aggregation-prone tail (448–511) was removed (Figure S2). The comparative crosslinking strategy yielded crosslinked peptides similar to those for the isolated ScR2TP complex and more importantly, crosslinked peptides of Nop58p to Pih1p and Rvb1p (Table S2). Nop58p_447 primarily crosslinked to Pih1p (Lys100, Lys393, and Lys411 of Nop58p to Lys52, Lys199, and Lys233 of Pih1p) and, to a lesser extent, to Rvb1p (Ser140 of Nop58p to Lys234 of Rvb1p) (Figures 7A and S6). Therefore, Pih1p serves as the adaptor for Nop58p_447 and places Nop58p_447 on top of the ScR2TP complex. We also noted some changes in crosslinked peptides between Pih1p and Rvb1/2p but not between Rvb1p and Rvb2p (Table S2) from those in the absence of Nop58p_447, suggesting possible conformational changes within Pih1pupon Nop58p_447 binding.
Sedimentation velocity experiments further support this architecture. Incubated Nop58p_447 and ScR2TP resulted in two major hydrodynamic species at 3.7 S and 13.7 S (Figure 7B). The 3.7 S species was assigned to free Nop58p_447 and Pih1p-Tah1p based on separately obtained sedimentation coefficient profiles in the absence of ScR2TP (Figure 7B). The slight increase in sedimentation coefficient of the larger species (13.7 S) from that of the ScR2TP hexamer (12.4 S) (Figure 2A) is likely due to the assembly of Nop58p_447 with ScR2TP in its hexameric form.
Placement of Nop58p on Pih1p that contacts the nucleotide-sensitive DII predicts a mechanism of its dissociation through nucleotide-induced conformation of DII in Rvb1/2p (Figure 7C).
DISCUSSION
Assembly of snoRNPs likely involves anchoring intermediates, masking non-productive assembly paths, stepwise folding of snoRNA and proteins, and finally releasing the assembly factors. Recent advances in genomics and proteomics have led to identification of several protein complexes required for (and acting at different stages of) box C/D snoRNP assembly (Boulon et al., 2008; Gonzales et al., 2005; King et al., 2001; Massenet et al., 2016; McKeegan et al., 2007, 2009; Quinternet et al., 2015; Rothe et al., 2014; Zhao et al., 2005, 2008). The molecular basis for these processes remains largely unknown. The ScR2TP complex is an essential snoRNP assembly factor that impacts on snoRNP stability and the nucleolus localization, which is required for ribosome maturation. Although the biological significance of ScR2TP in snoRNP assembly has been well established, its structural characterization lags behind. We now provide an initial description of the yeast ScR2TP complex and its interaction with snoRNP protein Nop58 through biophysical and cryo-EM analysis. We show that the ScR2TP complex is composed of a hexameric platform with a Pih1p-Tah1p cap that is likely the site of assembly of all snoRNP intermediates. The hexameric structure of the reconstituted ScR2TP complex is consistent with the observed hexamer of an endogenously purified ScR2TP complex (Cheung et al., 2010b) and suggests a structural mechanism for the nucleotide-driven chaperone activity (Figure 7C).
Despite a preference for dodecamers by free Rvb1/2p (Cheung et al., 2010a), Pih1p-Tah1p shifts it toward a hexameric assembly. This structural model of the assembled ScR2TP is reminiscent of that of other Rvb-associated complexes. EM studies of yeast endogenous INO8O and SWR-C complexes, both possessing Rvb1/2p and significantly larger in size, showed a similar architecture built from the single heterohexameric ring of Rvb1/2p with a flexible tail composed of other subunits (Nguyen et al., 2013; Tosi et al., 2013; Watanabe et al., 2015). The architectural similarity among the different Rvb1/2p complexes suggests a structural plasticity at the Rvb1/2p binding surface. Our unique observation that the IDR of Pih1p directly contacts the insertion domain of Rvb1/2p provides a structural basis for the plasticity. Pih1p IDR and the insertion domain can mutually adapt to each other, leading to an optimal binding interface. Our analytical ultracentrifugation result that Pih1p-Tah1p binding to Rvb1/2p competes for formation of free Rvb1/2p do-decamer supports the favorable interaction and highlights the importance of IDRs in Rvb1/2p-mediated assemblies.
Although it was noted previously that nucleotides prevented Pih1p-Tah1p from associating with Rvb1/2p (Kakihara et al., 2014), the molecular basis for this observation remained unclear. We demonstrated by EM imaging that nucleotides can release Pih1p-Tah1p heterodimer from the pre-formed ScR2TP complex. On the basis that Pih1p-Tah1p binds through the insertion domain of Rvb1/2p, our findings establish that the insertion domain, a structural element unique to Rvb1/2 among the AAA+ superfamily of proteins and a known sensor of nucleotide binding, therefore couples nucleotide binding to Pih1p-Tah1p dissociation. A possible mechanism would be simply rearranging the Pih1p-Tah1p binding site through conformational changes in the insertion domain by nucleotides. Taking this mechanism a step further, any client protein associated with ScR2TP through Pih1p-Tah1p could also be released as a result of nucleotide binding. Our interaction mapping of the box C/D protein Nop58p supports this client loading/release model (Figure 7C).
Our study did not address experimentally if and how Hsp90 participates in ScR2TP assembly or ScR2TP-Nop58p interaction but did shed some light on this process. Previous studies had showed convincingly that the three proteins, Hsp90, Pih1p, and Tah1p, form a complex (Pal et al., 2014; Zhao et al., 2008) that, however, does not co-purify with ScR2TP in yeast (Zhao et al., 2008). This suggests that Pih1p-Tah1p forms a distinct chaperone complex from R2TP with Hsp90. Our interaction mapping identified Pih1p as the adaptor for Nop58p, thereby making it possible for Nop58p to interact with both chaperone activities. Given the different localization sites between Hsp90 (cytosol) and Rvb1/2p (nucleoplasm), the two complexes may be required to meet the different needs in Nop58p maturation at two different locations.
Recently, another snoRNP “assembly complex” containing Rvbs, BCD1, Nufip, ZNHIT3, Nop58, and 15.5k was isolated in human (Bizarro et al., 2014). It is important to note that this complex lacks the human Pih1p-Tah1p complex homolog. However, it contains the 15.5k protein that is known to be the initiator protein for box C/D snoRNP assembly. Interestingly, unlike ScR2TP, the BCD1-associated complex requires the presence of nucleotides (Bizarro et al., 2014). The fact that both complexes interact with Nop58 that in turn binds the 15.5k-bound snoRNA suggests a nucleotide-driven snoRNP assembly process. Upon ATP binding and hydrolysis, R2TP can “hand over” Nop58 to the BCD1-associated complex for binding with the 15.5k-snoRNA complex in order to accomplish the initial assembly. The Rvb1/2p complex was also shown by yeast two-hybrid studies to interact with the homolog of Nop58, Prp31, a U4 small nuclear ribonucleoprotein (snRNP) component. Similarly, Prp31 binds to U4 snRNA following the 15.5k (human)/Snu13p (yeast) binding to U4. Therefore, the same principle may apply to U4 snRNP assembly as well. Understanding the dynamic process of snoRNP assembly through these seemingly complicated machines requires thorough characterization of their structural properties.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
KEY RESOURCE TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| Rosetta (DE3) | EMD Millipore (Novagene) | Cat# 70954 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Q5 site-specific mutagenesis kit | New England Biolab | Cat# E0554S |
| BS3-d0/d4 | Thermo Scientific | Cat# 21590/21595 |
| Glutaraldehyde solution | Sigma Aldrich | Cat# G7651–10ML |
| ProteoExtract all-in-one trypsin digestion kit | Calbiochem | Cat# 650212 |
| Uranyl Formate | Electron Microscopy Sciences | Cat# 22450 |
| Deposited Data | ||
| Experimental map | This paper | EMD-8839 |
| Recombinant DNA | ||
| pHRvb1 | This paper | |
| pRvb2 | This paper | |
| AII-Rvb2p | This paper | |
| pHPih1-Tah1 | This paper | |
| pHNop58_447 | This paper | |
| Software and Algorithms | ||
| UltraScanIII | (Demeler, 2010) | http://www.ultrascan3.uthscsa.edu/ |
| US-SOMO | (Brookes et al., 2010) | http://www.somo.uthscsa.edu/ |
| Zeno | (Mansfield et al., 2001) | https://doi.org/10.18434/T48W2J |
| Appion | (Lander et al., 2009) | http://www.appion.org |
| ACE | (Mallick et al., 2005) |
http://nramm.scripps.edu/software/ace http://graphics.ucsd.edu/spmallick/research/ace |
| CTFFIND | (Rohou and Grigorieff, 2015) | http://grigoriefflab.janelia.org/ctf |
| FindEM | (Roseman, 2004) | |
| Relion | (Scheres, 2012) | http://www2.mrc-lmb.cam.ac.uk/relion/index.php/Main_Page |
| Frealign | (Grigorieff, 2016) | http://grigoriefflab.janelia.org/frealign |
| ProteoWizard MSConvert | (Chambers et al., 2012) | http://proteowizard.sourceforge.net |
| Chimera | (Pettersen et al., 2004) | http://www.cgl.ucsf.edu/chimera/ |
| Other | ||
| Ni-NTA agrose | Qiagen | Cat# 30230 |
| Superdex 200 PC 3.2/30 | GE healthcare | Cat# 17–1089-01 |
| Hiload 16/60 superdex 200 | GE healthcare | Cat# 17–1069-01 |
| Superdex 200 Increase 10/300 GL | GE healthcare | Cat# 28–9909-44 |
| Mono Q 5/50 GL | GE healthcare | Cat# 17–5166-01 |
| R 2/2 200 mesh Cu holey carbon grids | Quantifoil | |
| Carbon-only 400 mesh grids Cu | Electron Microscopy Sciences | Cat# CF400-cu |
CONTACT FOR REAGENT AND RESOURCES SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Hong Li (hong.li@fsu.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Bacterial Strains Used for Protein Expression
Rosetta(DE3) Escherichia Coli cells (from Novagen) were cultured in Luria Broth (LB) with shaking at 37°C until 0D600 reached 0.6–0.8, supplemented with antibiotics. The cells continued to grow at 16°C overnight after being induced by 0.2mM isopropyl β-D-1-thiogalactopyranoside (IPTG).
METHOD DETAILS
Protein Expression and Purification
DNA sequences encoding the proteins were PCR-amplified from the Saccharomyces cerevisiae (S288c) genome. The gene expressing full-length Rvb1p with an N-terminal 6xHis tag followed by a TEV cleavage site was cloned into pET11b (pHRvb1) and that expressing the full-length Rvb2p was cloned into pET13b (pRvb2).The plasmids encoding Rvb2p without its Domain II (DII-Rvb2p) were constructed using the Q5 mutagenesis kit (New England Biolab). The genes expressing the N-terminal 6xHis Pih1p and non-tagged Tah1p were cloned into a single pqLink N vector for co-expression. Isolated His-Pih1p and its truncation mutants were expressed from a pqLink N vector containing the pih1 sequence fused with 6xHis codons. The gene expressing residues 1–447 of Nop58p with an N-terminal 6xHistag was cloned into pET11b (pHNop58_447).
Proteins were expressed in E. coli cells and purified with nickel affinity chromatography followed by size-exclusion chromatography. The Rvb1/2p complex was co-purified from E. coli cells cotransformed with both pHRvb1 and pRvb2. Cells were grown at 37°C until 0D600 reached 0.6–0.8 before being induced with 0.2 mM IPTG. The cells were allowed to grow at 16°C overnight before being harvested at 4 °C by pelleting at 6000 × g for 15 min in a Beckman Coulter Avanti J-20 XPI centrifuge (Optima). To purify the proteins, cell pellets were suspended in buffer A (25 mM sodium phosphate, pH 7.5 500 mM NaCl, and 5% glycerol) and lysed by sonication. The clear supernatant was then applied to a Ni-NTA column (Qiagen) equilibrated with buffer A. The bound sample was washed with buffer B (25 mM sodium phosphate, pH 7.5, 500 mM NaCl, 35 mM imidazole, and 5% glycerol) and eluted with buffer B supplemented with 350 mM imidazole. The Ni-NTA pool was directly applied to gel filtration and fractions were pooled before being flash-frozen for storage at −80° C. To obtain non-tagged Rvb1/2p, the Ni-NTA pool was digested with TEV protease for two h at room temperature. The cleavage mixture was dialyzed against buffer A and applied to a Ni-NTA column. The flow through was collected and buffer-exchanged to buffer C (25 mM HEPES pH 7.5 and 150 mM NaCl) by size exclusion chromatography. The Pih1p-Tah1p complex was purified similarly as Rvb1/2p complex by Ni-NTA affinity chromatography followed by size exclusion chromatography, except for that 100 mM NaCl and 10% glycerol was used in the Ni-affinity purification buffers.
Nop58p 1–447 fragment was purified by the similar method described above for the Rvb1/2p complex with the exception of an additional purification step with a MonoQ column after gel filtration. Pooled Nop58p fractions were dialyzed against buffer C and concentrated by an Amicon Ultra Centrifugal Filter device to 5 mg/ml before being flash-frozen for storage at −80°C.
To assemble the ScR2TP complex, purified Pih1p-Tah1p and non-tagged Rvb1/2p were mixed and applied to a Ni-affinity column in buffer A. The eluate was loaded onto a Superdex S200 column in buffer C, and fractions corresponding to the ScR2TP complex were taken directly for EM analysis. The R22TP complex was purified similarly except that the fractions corresponding to the complex were pooled and concentrated before the Grafix procedure (Stark, 2010).
Analytical Gel Filtration
Purified Rvb1/2p and Pih1p-Tah1p complexes were mixed in a 1:4 molar ratio in buffer C and incubated at 4°C overnight. Rvb1/2p and Pih1p-Tah1p were also incubated separately as controls under the same conditions. The protein samples were applied to a Superdex 200 3.2/300 column. Fractions were collected and analyzed by silver-stained SDS-PAGE.
Isothermal Titration Calorimetry
Titration experiments were performed by isothermal titration calorimetry (ITC) using a VP-ITC microcalorimeter from Microcal, Inc. (Northampton, MA), interfaced with a computer. The titration calorimeter consists of 1.45 ml sample cell containing a protein solution and a matched thermal reference cell filled with the same buffer. Pih1p-Tahp1 and Rvb1/2p complexes were prepared with the same buffer by gel filtration. Prior to the experiment, both samples were filtered and degassed under vacuum for 10 min in a thermo Vac system (MicorCal). Rvb1/2p was placed into the sample cell at 1.8 μM and Pih1p-Tah1p at 40-fold excess was loaded into the injection needle. Titrations consisted of 40× 6μl injections, with at least 200 sec intervals. All runs were made at constant stirrer speed of 310 rpm, and all experiments were performed at 25°C. The evolved heat peaks were integrated and then fitted to a theoretical titration curve of a serial binding site model by non-linear least squares to yield thermodynamic parameters. All isotherms show negative deflection indicative of an exothermic reaction. A separate run with Pih1p-Tah1p titrating into buffer showed insignificant endo-thermic, and equal sized dilution peaks, suggesting that at this concentration no aggregation of Pih1p-Tah1p (Figure 2B). The heat due to the dilution of Pih1p-Tah1p was subtracted from the experimental data conducted with Rvb1/2p.
Fitting binding isotherms with the available models as implemented in the MicroCal Origin software revealed clearly a two-site serial binding model in which a tight-binding is followed by a weak binding site. The enthalpy change (ΔH0) and binding dissociation constants (K1d and K2d) were directly obtainable from the fitting. The entropy change-TΔS0 for both sites were subsequently calculated Kd and ΔH0 values (Figure 2B).
Analytical Ultracentrifugation (AUC)
The Rvb1/2pand Pih1p-Tah1p (or Pih1p truncation mutant) complexes were mixed at a 1:4 molar ratio in an AUC-compatible buffer (20 mM PBS pH 7.5,100 mM NaCl). The mixtures were incubated at 4°C overnight before loading into AUC cells. The Rvb1/2p concentration was held at 0.3 mg/ml in all runs. Sedimentation velocity experiments were performed at 20°C in a Beckman Optima XL-I analytical ultracentrifuge with an An60ti rotor. The sample (430 μL) was loaded into an Epon 2-channel standard cell, and a similar amount of water (450 μL) was loaded as reference. The rotor speed was 40,000 rpm for all the runs. Scans were acquired in intensity mode until sedimentation was completed.
Data analysis was performed by use of the UltraScan III software version 3.3 as described elsewhere (Demeler, 2010). Data optimization was achieved with two dimensional spectrum analysis (2DSA) (Brookes et al., 2010) as well as time-invariant and radially-invariant noise removal (Schuck and Demeler, 1999). Parametrically constrained spectrum analysis using a decreasing sigmoid parameterization, combined with Monte Carlo (Demeler and Brookes, 2008) was used to determine sedimentation coefficient distributions. Atomic structure models of the Sc Rvb1/2p created from Ct Rvb1/2 crystal structures were processed with US-SOMO (Brookes et al., 2010) to create bead models with overlaps. These models were further analyzed with Zeno (Mansfield et al., 2001) to derive hydrodynamic predictions.
Electron Microscopy Sample Preparation
To prepare samples for negative stain electron microscopy, 3 μL of the ScR2TP complex was applied onto continuous carbon-only 400 mesh grids (Electron Microscopy Sciences) that had been plasma-cleaned for 8 s by use of a Gatan SOLARIS model 950 advanced plasma cleaner, and negatively stained with 2% (w/v) uranyl formate. For cryoEM samples, 3 μL of 0.5 mg/ml ScR2TP was applied onto plasmid-cleaned 200 mesh Quantifoil® (Jena, Germany) R2/2 grids. The grids were vitrified by use of an FEI vitrobot (Hillsboro OR) by blotting for 3 s before being plunged into liquid ethane.
Purified R22TP complex was prepared by fractionation with GraFix (Kastner et al., 2008) with a glycerol gradient from 10% to 30% containing 0.2% glutaraldehyde. 200 μL sample at 6 mg/mL was loaded onto the top of the centrifuge tube. The sample was centrifuged at 33,000 rpm for 17 h at 4°C in an sw40ti rotor in a Beckman Coulter XL-100K centrifuge (Optima). After centrifugation, the gradient was fractioned into 24 0.5 mL fractions by pipetting. Fractions were examined with negative stain EM by use of a CM120BT microscope and the most homogenous fraction was selected for further data collection. Glycerol gradient-fractionated and crosslinked R22TP complex was placed onto a glow-discharged continuous carbon grid and stained with 2% uranyl formate.
EM Data Collection and Processing
The negatively stained R22TP, ScR2TP, and the ice-embedded ScR2TP samples were imaged with an FEI Titan Krios operated at 300 keV equipped with a DE 20 direct electron camera. The Leginon software for automatic data acquisition was used (Suloway et al., 2005). A total of 2052 images of negatively stained R22TP and 593 images of negatively stained ScR2TP were collected at 22,500 x magnification with 1.62 A/pixel sampling rate at the specimen level and −1.0 to −3.0 microns defocus. For cryoEM of ScR2TP, 1396 images were collected at 29,000x magnification with 1.22 Å/pixel sampling rate and a total dose of 58.68 e-/Å2. Frames from the DE 20 were aligned and dose compensated using the alignment script provided by DE as implemented in Appion (Spear et al., 2015).
Images were processed by use of Appion (Lander et al., 2009). CTFs were initially estimated with the Automated CTF Estimation (ACE) (Mallick et al., 2005) software package and refined by use of CTFFIND (Rohou and Grigorieff, 2015). Images with an estimation confidence value of less than 0.7 were rejected. Particles were initially picked manually and aligned in a reference-free manner with topology alignment (Ogura et al., 2003) to create 2D templates for template-based picking. Particles were then automatically picked using FindEM (Roseman, 2004). This procedure resulted in 38,266 particles for the R22TP complex, 24,088 particles for negatively stained ScR2TP, and 81,089 particles for the cryo ScR2TP complex. For all data sets, reference-free alignment and classification were performed by use of topology alignment to reject bad picks, aggregates, and poorly aligned particles. This resulted in 35,489 particles for the R22TP complex, 17,685 for negatively stained ScR2TP, and 54,712 for the cryo ScR2TP. For all data sets, particles were initially reconstructed with Relion refinement 3D classification (Scheres, 2012). The initial model for R22TP was from the crystal structure of Ct Rvb1/2p complex (PDB: 4WW4) phase randomized and low-pass filtered to 50 Å. The initial models for the ScR2TP data sets were the larger R22TP structure (Figure S7) similarly filtered to 60 Å. Heterogeneity in samples with the bound Pih1p-Tah1p led to misclassification of particles, limiting the resolution of the reconstructions (Figure S7). For all data sets, particles that resulted in a reconstruction with discernable features were subjected to further refinement with Frealign either with C1 symmetry (for R22TP) or pseudoC3 symmetry (for ScR2TP). The pseudoC3 option in Frealign minimizes misalignment of the asymmetric parts due to their correlation with symmetrically related orientations by testing the final model against particle orientations (Grigorieff, 2016). All EM density maps were visualized by use of UCSF Chimera (Pettersen et al., 2004).
Lysine-Specific Cross-Linking Coupled With Mass Spectrometry
The ScR2TP complex analyzed by cryoEM or its complex with Nop58p at an 1:4 ScR2TP:Nop58p molar ratio was incubated with 0.5 % deuterated (d4) or nondeuterated (d0) bis(sulfosuccinimidyl)suberate (BS3) for 30 min. The BS3-d0 and BS3-d4 crosslinked samples were mixed in equal volume and loaded onto a 7.5 % SDS-PAGE gel. The band corresponding to the crosslinked protein complex was excised from the gel, and in-gel digested with the ProteoExtract all-in-one trypsin digestion kit (catalog # 650212, Calbiochem, Billerica, MA) following the manufacture recommended protocol. Briefly, the gel band was destained with a wash buffer, cut into ~ 3 mm pieces, shrunk with acetonitrile and dried. Gel pieces were then rehydrated with a digest buffer and reduced with a reducing agent at 37°Cfor 10 min followed by cooling to room temperature and treatment with a blocking reagent for 10 min. Trypsin was added to the mixture and the reaction was incubated at the room temperature overnight. The supernatant was collected after centrifugation and dried before performing mass spectrometry.
The dried peptide mixture was re-dissolved in H20 containing 0.5 % formic acid and separated by nLC by use of an Easy Nano LC II system (Thermo Fisher Scientific) with a 100 mm x 2 cm trap column (easy column, catalog # SC001 Thermo Fisher Scientific) and a 75 μm × 10 cm C18AQ analytical column (Thermo Fisher Scientific). Mobile phases were A (99.9 % H20 and 0.1% formic acid) and B (99.9 % acetonitrile and 0.1% formic acid). A linear gradient from 1% to 45% B over 3 hours was performed at a flow rate of 300 nL/min. Eluate was on-line ionized by electrospray ionization (ESI) and detected by a Velos LTQ-Orbitrap Mass Spectrometer (Thermo Scientific). The precursor ions were detected at a software-defined mass resolving power of 60 K (at m/z 800) in the Orbitrap. Data-dependent MS2 was performed for the top 10 most abundant precursor ions with collision induced dissociation (CID) in the LTQ. The .raw file was converted to an .mzXML file by an open-access software ProteoWizard MSConvert (Chambers et al., 2012). The .mzXML file was then analyzed by the open-access software StavroX for identification of crosslinked peptides by comparing measured precursor and fragment ion masses to the theoretical masses of BS3-d0 crosslinked peptides generated in silico (Gotze et al., 2012). The following parameters were used in StavroX: up to 3 trypsin miscleavages; static modification of cysteine to cysteine acetamide; variable modification of methionine by oxidation; precursor precision of < 3 ppm; fragment ion precision of < 0.8 Da; S/N ratio higher than 2. StavroX results were then manually checked to verify crosslinked peptides that show the presence of both BS3-d0 and BS3-d4 crosslinkers, ion pairs separated by 4.0247 Da. Validated corss-linked peptides were checked again for peptide sequence assignment by comparing measured b and y fragment ion masses to their theoretical values.
Stoichiometry and Protein Identification by Mass Spectrometry
Stoichiometry of the ScR2TP complex was established by label-free quantitative mass spectrometry as previously described (Smits and Vermeulen, 2013). Trypsin-digested peptides from copurified ScR2TP were prepared similarly as above and applied to online nanoLC-MS/MS. LC-MS/MS data was acquired with an LTQ Orbitrap Velos (Thermo Fisher Scientific) and processed with MaxQuant software at standard settings. Relative stoichiometry of the four proteins was obtained by dividing the background corrected magnitude-based absolute quantification (iBAQ) values of the target protein by those of protein standards. To identify the presence of components, the GraFix separated R22TP was exchanged into a mass spectrometry-compatible buffer (25 mM (NH4)2CO3, pH 8.0) and was digested with trypsin followed by treatment with 3% acetonitrile in LC-MS grade water (J.T. Baker) with 0.1 % formic acid. Peptide masses were analyzed by MALDI-TOF with anXevoTQ-S UPLC-nESI-triplequadrupole (Waters), coupled to a Synapt G2 HDMS (Waters). Rabbit alcohol dehydrogenase 1 served as an internal control. Raw data were generated by MassLynx (Waters) followed by processing with ProteinLynx Globle SEVER (Waters).
Hydrogen Deuterium Exchange (HDX)
Purified Pih1p-Tah1p at 20 μM in 150 μM NaCl and 20 mM HEPES, pH 7.5, or the same sample mixed with separately purified Rvb1/2p at 30 μM in Buffer C were used for HDX analysis. Seven 5 μL aliquots of either Pih1p-Tah1p alone or its complex with Rvb1/2p were mixed with 45 μL of a D2O buffer to initiate HDX. At the end of incubation periods of 0.5, 1, 4, 15, 30, 60, and 480 min for each reaction, the sample was quenched by mixing with 25 μL of 8 M urea and 200 mM TCEP in 1 % formic acid and digested by Protease XIII for 3 min at 0.4°C as described previously (Zhang et al., 2013). Each HDX sample was then subjected to on-line LC-MS. The ionized eluate was directed to a custom-built Velso Pro (Thermo Scientific) 21 T FT-ICR mass spectrometer (Hendrickson et al., 2015). The instrument was externally calibrated with peptide standards. The target ion number was set to three million by a single fill of ions with automatic gain control. Time-domain ICR data were collected for 0.76 seconds, Hanning apodized and zero-filled once before discrete Fourier transformation. Mass spectra were collected from 400 < m/z < 1,300 at a high mass resolving power (m/Δm50% = 200,000 at m/z 400, in which Δm50% is magnitude mass spectral peak full width at half-maximum peak height). Data was acquired by the use of Xcalibur (Thermo Fisher Scientific) and analyzed by a custom analysis package as described previously (Blakney et al., 2011; Kazazic et al., 2010).
DATA AVAILABILITY
The R2TP structure has been deposited in the EM Data Bank (EMDB) under EMDB code 8839.
Supplementary Material
Highlights.
The yeast R2TP chaperone complex forms a dome-shaped architecture
Rvb1/2p is the base and one Pih1p-Tah1 p forms the top of the dome
A client protein, Nop58, interacts with R2TP through binding to Pih1 p on top
The nucleotide-sensitive insertion domain of Rvb1/2p drives client release
ACKNOWLEDGMENTS
We thank John Spear for assistance in EM data collection, Shelby Davis for plasmid preparation, and B. Washburn and C. Pye of the Molecular Cloning Facility at FSU for the cloning experiments. This work was supported by NIH grants R01 GM66958 and R01 GM099604 to H.L. and NSF Division of Materials Research through DMR-11–57490 to A.G.M. The Ultra Scan development is supported by National Science Foundation grant ACI-1339649 (to B.D.), and AUC data analysis is supported by XSEDE allocation grant MCB-070039 (to B.D.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures, two tables, and one movie and can be found with this article online at http://dx.doi.org/10.1016Zj.str.2017.08.002.
REFERENCES
- Back R, Dominguez C, Rothe B, Bobo C, Beaufils C, Morera S, Meyer P, Charpentier B, Branlant C, Allain FH, et al. (2013). High-resolution structural analysis shows how Tah1 tethers Hsp90 to the R2TP complex. Structure 21, 1834–1847. [DOI] [PubMed] [Google Scholar]
- Bizarro J, Charron C, Boulon S, Westman B, Pradet-Balade B, Vandermoere F, Chagot ME, Hallais M, Ahmad Y, Leonhardt H, et al. (2014). Proteomic and 3D structure analyses highlight the C/D box snoRNP assembly mechanism and its control. J. Cell Biol. 207, 463–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizarro J, Dodre M, Huttin A, Charpentier B, Schlotter F, Branlant C, Verheggen C, Massenet S, and Bertrand E (2015). NUFIP and the HSP90/R2TP chaperone bind the SMN complex and facilitate assembly of U4-specific proteins. Nucleic Acids Res. 43, 8973–8989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakney GT, Hendrickson CL, and Marshall AG (2011). Predator data station: a fast data acquisition system for advanced FT-ICR MS experiments. Int. J. Mass Spectrom. 306, 246–252. [Google Scholar]
- Boulon S, Marmier-Gourrier N, Pradet-Balade B, Wurth L, Verheggen C, Jady BE, Rothe B, Pescia C, Robert MC, Kiss T, et al. (2008). The Hsp90 chaperone controls the biogenesis of L7Ae RNPs through conserved machinery. J. Cell Biol. 180, 579–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S, Pradet-Balade B, Verheggen C, Molle D, Boireau S, Georgieva M, Azzag K, Robert MC, Ahmad Y, Neel H, et al. (2010). HSP90 and its R2TP/Prefoldin-like cochaperone are involved in the cyto-plasmic assembly of RNA polymerase II. Mol. Cell 39, 912–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes E, Demeler B, Rosano C, and Rocco M (2010). The implementation of SOMO (SOlution Modeller) in the UltraScan analytical ultracentrifugation data analysis suite: enhanced capabilities allow the reliable hydrodynamic modeling of virtually any kind of biomacromolecule. Eur. Biophys. J. 39, 423–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaker-Margot M, Barandun J, Hunziker M, and Klinge S (2017). Architecture of the yeast small subunit processome. Science 355, eaal1880. [DOI] [PubMed] [Google Scholar]
- Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, Gatto L, Fischer B, Pratt B, Egertson J, et al. (2012). A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30, 918–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung KL, Huen J, Houry WA, and Ortega J (2010a). Comparison of the multiple oligomeric structures observed for the Rvb1 and Rvb2 proteins. Biochem. Cell Biol. 88, 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung KL, Huen J, Kakihara Y, Houry WA, and Ortega J (2010b). Alternative oligomeric states of the yeast Rvb1/Rvb2 complex induced by histidine tags. J. Mol. Biol. 404, 478–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeler B (2010). Methods for the design and analysis of sedimentation velocity and sedimentation equilibrium experiments with proteins. Curr. Protoc. Protein Sci. Chapter 7. Unit 7 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeler B, and Brookes E (2008). Monte Carlo analysis of sedimentation experiments. Colloid Polym. Sci. 286, 129–137. [Google Scholar]
- DiMaio F, Song Y, Li X, Brunner MJ, Xu C, Conticello V, Egelman E, Marlovits TC, Cheng Y, and Baker D (2015). Atomic-accuracy models from 4.5-A cryo-electron microscopy data with density-guided iterative local refinement. Nat. Methods 12, 361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewens CA, Su M, Zhao L, Nano N, Houry WA, and Southworth DR (2016). Architecture and nucleotide-dependent conformational changes of the Rvb1-Rvb2 AAA+ complex revealed by cryoelectron microscopy. Structure 24, 657–666. [DOI] [PubMed] [Google Scholar]
- Forget D, Lacombe AA, Cloutier P, Al-Khoury R, Bouchard A, Lavallee-Adam M, Faubert D, Jeronimo C, Blanchette M, and Coulombe B (2010). The protein interaction network of the human transcription machinery reveals a role for the conserved GTPase RPAP4/GPN1 and microtubule assembly in nuclear import and biogenesis of RNA polymerase II. Mol. Cell. Proteomics 9, 2827–2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia De La Torre J, Huertas ML, and Carrasco B. (2000). Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophys. J. 78, 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales FA, Zanchin NI, Luz JS, and Oliveira CC (2005). Characterization of Saccharomyces cerevisiae Nop17p, a novel Nop58p-inter-acting protein that is involved in Pre-rRNA processing. J. Mol. Biol. 346, 437–455. [DOI] [PubMed] [Google Scholar]
- Gotze M, Pettelkau J, Schaks S, Bosse K, Ihling CH, Krauth F, Fritzsche R, Kuhn U, and Sinz A (2012). StavroX—a software for analyzing crosslinked products in protein interaction studies. J. Am. Soc. Mass Spectrom. 23, 76–87. [DOI] [PubMed] [Google Scholar]
- Gribun A, Cheung KL, Huen J, Ortega J, and Houry WA (2008). Yeast Rvb1 and Rvb2 are ATP-dependent DNA helicases that form a heterohexameric complex. J. Mol. Biol. 376, 1320–1333. [DOI] [PubMed] [Google Scholar]
- Grigorieff N (2016). Frealign: an exploratory tool for single-particle cryo-EM. Methods Enzymol. 579, 191–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson CL, Quinn JP, Kaiser NK, Smith DF, Blakney GT, Chen T, Marshall AG, Weisbrod CR, and Beu SC (2015). 21 Tesla Fourier transform ion cyclotron resonance mass spectrometer: a national resource for ultrahigh resolution mass analysis. J. Am. Soc. Mass Spectrom. 26, 1626–1632. [DOI] [PubMed] [Google Scholar]
- Horejsi Z, Takai H, Adelman CA, Collis SJ, Flynn H, Maslen S, Skehel JM, de Lange T, and Boulton SJ (2010). CK2 phospho-dependent binding of R2TP complex to TEL2 is essential for mTOR and SMG1 stability. Mol. Cell 39, 839–850. [DOI] [PubMed] [Google Scholar]
- Huen J, Kakihara Y, Ugwu F, Cheung KL, Ortega J, and Houry WA (2010). Rvb1-Rvb2: essential ATP-dependent helicases for critical complexes. Biochem. Cell Biol. 88, 29–40. [DOI] [PubMed] [Google Scholar]
- Izumi N, Yamashita A, Iwamatsu A, Kurata R, Nakamura H, Saari B, Hirano H, Anderson P, and Ohno S (2010). AAA+ proteins RUVBL1 and RUVBL2 coordinate PIKK activity and function in nonsense-mediated mRNA decay. Sci. Signal. 3, ra27. [DOI] [PubMed] [Google Scholar]
- Jeganathan A, Leong V, Zhao L, Huen J, Nano N, Houry WA, and Ortega J (2015). Yeast rvb1 and rvb2 proteins oligomerize as a conformationally variable dodecamer with low frequency. J. Mol. Biol. 427, 1875–1886. [DOI] [PubMed] [Google Scholar]
- Jha S, and Dutta A (2009). RVB1/RVB2: running rings around molecular biology. Mol. Cell 34, 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakihara Y, and Houry WA (2012). The R2TP complex: discovery and functions. Biochim. Biophys. Acta 1823, 101–107. [DOI] [PubMed] [Google Scholar]
- Kakihara Y, Makhnevych T, Zhao L, Tang W, and Houry WA (2014). Nutritional status modulates box C/D snoRNP biogenesis by regulated subcellular relocalization of the R2TP complex. Genome Biol. 15, 404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastner B, Fischer N, Golas MM, Sander B, Dube P, Boehringer D, Hartmuth K, Deckert J, Hauer F, Wolf E, et al. (2008). GraFix: sample preparation for single-particle electron cryomicroscopy. Nat. Methods 5, 53–55. [DOI] [PubMed] [Google Scholar]
- Kazazic S, Zhang HM, Schaub TM, Emmett MR, Hendrickson CL, Blakney GT, and Marshall AG (2010). Automated data reduction for hydrogen/deuterium exchange experiments, enabled by high-resolution Fourier transform ion cyclotron resonance mass spectrometry. J. Am. Soc. Mass Spectrom. 21, 550–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King TH, Decatur WA, Bertrand E, Maxwell ES, and Fournier MJ (2001). A well-connected and conserved nucleoplasmic helicase is required for production of box C/D and H/ACA snoRNAs and localization of snoRNP proteins. Mol. Cell. Biol. 21, 7731–7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornprobst M, Turk M, Kellner N, Cheng J, Flemming D, Kos-Braun I, Kos M, Thoms M, Berninghausen O, Beckmann R, et al. (2016). Architecture of the 90S pre-ribosome: a structural view on the birth of the eukaryotic ribosome. Cell 166, 380–393. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Keogh MC, Datta N, Sawa C, Ryan OW, Ding H, Haw RA, Pootoolal J, Tong A, Canadien V, et al. (2003). A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 12, 1565–1576. [DOI] [PubMed] [Google Scholar]
- Lakomek K, Stoehr G, Tosi A, Schmailzl M, and Hopfner KP (2015). Structural basis for dodecameric assembly states and conformational plasticity of the full-length AAA+ ATPases Rvb1 Rvb2. Structure 23, 483–95. [DOI] [PubMed] [Google Scholar]
- Lakshminarasimhan M, Boanca G, Banks CA, Hattem GL, Gabriel AE, Groppe BD, Smoyer C, Malanowski KE, Peak A, Florens L, et al. (2016). Proteomic and genomic analyses of the Rvb1 and Rvb2 interaction network upon deletion of R2TP complex components. Mol. Cell. Proteomics 15, 960–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, et al. (2009). Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 166, 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang B, and Li H (2011). Structures of ribonucleoprotein particle modification enzymes. Q. Rev. Biophys. 44, 95–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado-Pinilla R, Liger D, Leulliot N, and Meier UT (2012). Mechanism of the AAA+ ATPases pontin and reptin in the biogenesis of H/ACA RNPs. RNA 18, 1833–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick SP, Carragher B, Potter CS, and Kriegman DJ (2005). ACE: automated CTF estimation. Ultramicroscopy 104, 8–29. [DOI] [PubMed] [Google Scholar]
- Mansfield ML, Douglas JF, and Garboczi EJ (2001). Intrinsic viscosity and the electrical polarizability of arbitrarily shaped objects. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 64, 061401. [DOI] [PubMed] [Google Scholar]
- Massenet S, Bertrand E, and Verheggen C (2016). Assembly and trafficking of box C/D and H/ACA snoRNPs. RNA Biol. 74, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matias PM, Gorynia S, Donner P, and Carrondo MA (2006). Crystal structure of the human AAA+ protein RuvBL1. J. Biol. Chem. 281, 38918–38929. [DOI] [PubMed] [Google Scholar]
- McKeegan KS, Debieux CM, Boulon S, Bertrand E, and Watkins NJ (2007). A dynamic scaffold of pre-snoRNP factors facilitates human box C/D snoRNP assembly. Mol. Cell. Biol. 27, 6782–6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeegan KS, Debieux CM, and Watkins NJ (2009). Evidence that the AAA+ proteins TIP48 and TIP49 bridge interactions between 15.5K and the related NOP56 and NOP58 proteins during box C/D snoRNP biogenesis. Mol. Cell. Biol. 29, 4971–4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nano N, and Houry WA (2013). Chaperone-like activity of the AAA+ proteins Rvb1 and Rvb2 in the assembly of various complexes. Philos. Trans. R. Soc. Lon. B Biol. Sci. 368, 20110399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman DR, Kuhn JF, Shanab GM, and Maxwell ES (2000). Box C/D snoRNA-associated proteins: two pairs of evolutionarily ancient proteins and possible links to replication and transcription. RNA 6, 861–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen VQ, Ranjan A, Stengel F, Wei D, Aebersold R, Wu C, and Leschziner AE (2013). Molecular architecture of the ATP-dependent chromatin-remodeling complex SWR1. Cell 154, 1220–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewiarowski A, Bradley AS, Gor J, McKay AR, Perkins SJ, and Tsaneva IR (2010). Oligomeric assembly and interactions within the human RuvB-like RuvBL1 and RuvBL2 complexes. Biochem. J. 429, 113–125. [DOI] [PubMed] [Google Scholar]
- Ogura T, Iwasaki K, and Sato C (2003). Topology representing network enables highly accurate classification of protein images taken by cryo electron-microscope without masking. J. Struct. Biol. 143, 185–200. [DOI] [PubMed] [Google Scholar]
- Paci A, Liu XH, Huang H, Lim A, Houry WA, and Zhao R (2012). The stability of the small nucleolar ribonucleoprotein (snoRNP) assembly protein Pih1 in Saccharomyces cerevisiae is modulated by its C terminus. J. Biol. Chem. 287, 43205–43214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal M, Morgan M, Phelps SE, Roe SM, Parry-Morris S, Downs JA, Polier S, Pearl LH, and Prodromou C (2014). Structural basis for phosphorylation-dependent recruitment of Tel2 to Hsp90 by Pih1. Structure 22, 805–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Prieto MB, Georg RC, Gonzales-Zubiate FA, Luz JS, and Oliveira CC (2015). Nop17 is a key R2TPfactorforthe assembly and maturation of box C/D snoRNP complex. BMC Mol. Biol. 16, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam CD, Clancy SB, Tsuruta H, Gonzalez S, Wetmur JG, and Tainer JA (2001). Structure and mechanism of the RuvB Holliday junction branch migration motor. J. Mol. Biol. 311, 297–310. [DOI] [PubMed] [Google Scholar]
- Quinternet M, Rothe B, Barbier M, Bobo C, Saliou JM, Jacquemin C, Back R, Chagot ME, Cianferani S, Meyer P, et al. (2015). Structure/function analysis of protein-protein interactions developed by the yeast Pih1 platform protein and its partners in box C/D snoRNP assembly. J. Mol. Biol. 427, 2816–2839. [DOI] [PubMed] [Google Scholar]
- Rohou A, and Grigorieff N (2015). CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman AM (2004). FindEM—a fast, efficient program for automatic selection of particles from electron micrographs. J. Struct. Biol. 145, 91–99. [DOI] [PubMed] [Google Scholar]
- Rothe B, Saliou JM, Quinternet M, Back R, Tiotiu D, Jacquemin C, Loegler C, Schlotter F, Pena V, Eckert K, et al. (2014). Protein Hit1, a novel box C/D snoRNP assembly factor, controls cellular concentration of the scaffolding protein Rsa1 by direct interaction. Nucleic Acids Res. 42, 10731–10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottbauer W, Saurin AJ, Lickert H, Shen X, Burns CG, Wo ZG, Kemler R, Kingston R, Wu C, and Fishman M (2002). Reptin and pontin antagonistically regulate heart growth in zebrafish embryos. Cell 111, 661–672. [DOI] [PubMed] [Google Scholar]
- Scheres SH (2012). RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C, and Robinson CV (2014). A comparative cross-linking strategy to probe conformational changes in protein complexes. Nat. Protoc. 9, 2224–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P, and Demeler B (1999). Direct sedimentation analysis of interference optical data in analytical ultracentrifugation. Biophys. J. 76, 2288–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Mizuguchi G, Hamiche A, and Wu C (2000). A chromatin remodelling complex involved in transcription and DNA processing. Nature 406, 541–544. [DOI] [PubMed] [Google Scholar]
- Smits AH, and Vermeulen M (2013). Stoichiometry determination of protein complexes by label-free quantitative mass spectrometry-based proteomics (Prot 61). Nucleic Acids Res. 41, e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear JM, Noble AJ, Xie Q, Sousa DR, Chapman MS, and Stagg SM (2015). The influence of frame alignment with dose compensation on the quality of single particle reconstructions. J. Struct. Biol. 192, 196–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark H (2010). GraFix: stabilization of fragile macromolecular complexes for single particle cryo-EM. Methods Enzymol. 481, 109–126. [DOI] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, and Carragher B (2005). Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60. [DOI] [PubMed] [Google Scholar]
- Sun Q, Zhu X, Qi J, An W, Lan P, Tan D, Chen R, Wang B, Zheng S, Zhang C, et al. (2017). Molecular architecture of the 90S small subunit preribosome. Elife 6, e22086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H, Xie Y, de Lange T, and Pavletich NP (2010). Tel2 structure and function in the Hsp90-dependent maturation of mTOR and ATR complexes. Genes Dev. 24, 2019–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosi A, Haas C, Herzog F, Gilmozzi A, Berninghausen O, Ungewickell C, Gerhold CB, Lakomek K, Aebersold R, Beckmann R, et al. (2013). Structure and subunit topology of the INO80 chromatin remodeler and its nucleosome complex. Cell 154, 1207–1219. [DOI] [PubMed] [Google Scholar]
- Tucker PA, and Sallai L (2007). The AAA+ superfamily—a myriad of motions. Curr. Opin. Struct. Biol. 17, 641–652. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Tan D, Lakshminarasimhan M, Washburn MP, Hong EJ, Walz T, and Peterson CL (2015). Structural analyses of the chromatin remodelling enzymes INO80-C and SWR-C. Nat. Commun. 6, 7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Chen J, Kuwajima K, Zhang HM, Xian F, Young NL, and Marshall AG (2013). Nucleotide-induced conformational changes of tetra-decameric GroEL mapped by H/D exchange monitored by FT-ICR mass spectrometry. Sci. Rep. 3, 1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Davey M, Hsu YC, Kaplanek P, Tong A, Parsons AB, Krogan N, Cagney G, Mai D, Greenblatt J, et al. (2005). Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell 120, 715–727. [DOI] [PubMed] [Google Scholar]
- Zhao R, Kakihara Y, Gribun A, Huen J, Yang G, Khanna M, Costanzo M, Brost RL, Boone C, Hughes TR, et al. (2008). Molecular chaperone Hsp90 stabilizes Pih1/Nop17 to maintain R2TP complex activity that regulates snoRNA accumulation. J. Cell Biol. 180, 563–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The R2TP structure has been deposited in the EM Data Bank (EMDB) under EMDB code 8839.


