Figure 1. Schematic Representation of ScR2TP Protein Features.
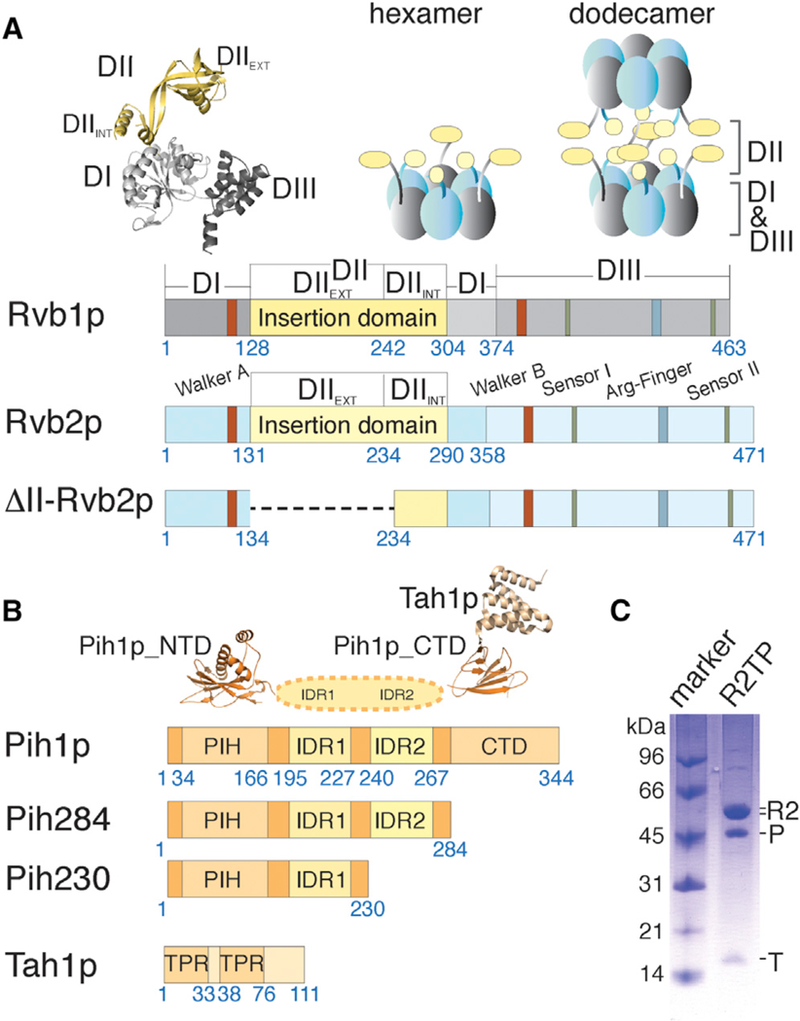
(A) Schematic diagrams depicting primary structural features of yeast Rvb1p and Rvb2p including both wild-type and mutant constructs used in this study. Residue numbers at the beginning and ends of each region discussed in the text are indicated under the diagrams. The crystal structure of human RUVBL1 (PDB: 1C9O) shown in ribbons illustrates the three conserved domains for both Rvb proteins colored as in the diagrams. Schematic arrangements of previously observed oligomers, heterohexamer and heterododecamer, for the yeast Rvb1/2p complex are illustrated above the diagrams by solid blocks in gray and light blue.
(B) Schematic diagrams depicting primary structural features of yeast Pih1p and Tah1p. Residue numbers at the beginning and ends of each region discussed in the text are indicated under the bar diagrams. The available crystal structures for the N-terminal domain of Pih1p (Pih1p_NTD) (PDB: 4CHH) and Tah1 p bound to the C-terminal domain of Pih1p(Pih1p_CTD) (PDB: 4CGU) are shown above the diagrams.
(C) Co-purified Saccharomyces cerevisiae ScR2TP complex from recombinantly expressed components is shown on an SDS-PAGE gel with a molecular weight marker. “R2” denotes the co-purified, tag-free Rvb1/2p complex (see a zoomed view in Figure S2), “P” denotes His-tagged Pih1p, and “T” denotes tag-free Tah1p. The stoichiometry of the co-purified ScR2TP was determined by label-free mass spectrometry to be ~0.5:1.6:3:3 (Table S1).
