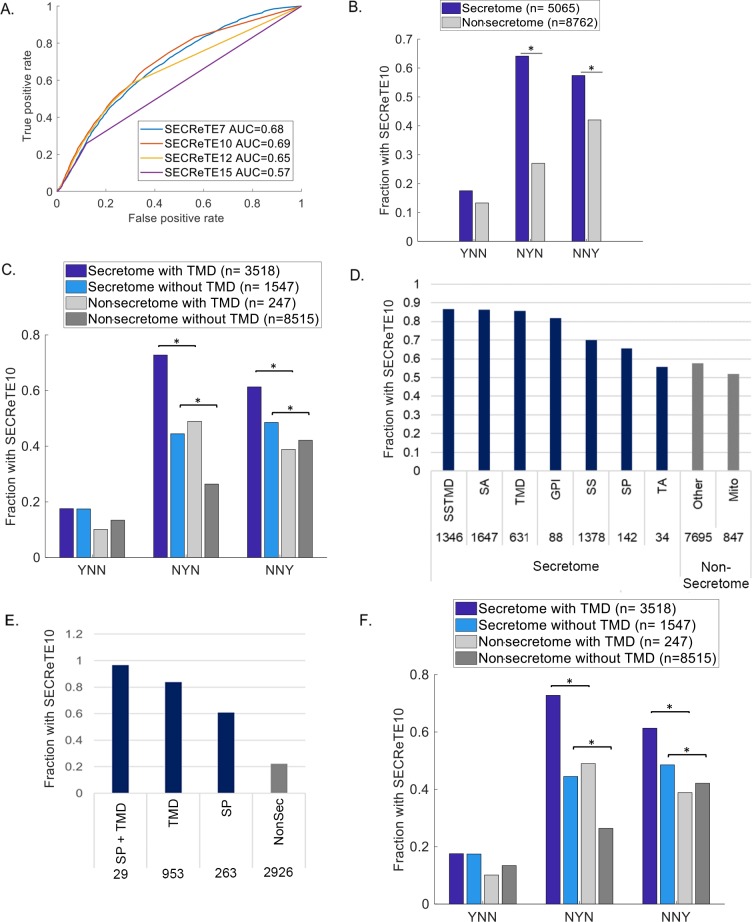
Fig 4. SECReTE enrichment in the secretomes of yeast, bacteria, and humans.
(A) SECReTE10 maximizes the ability to classify secretome genes in human. ROC curves were plotted for each of the indicated thresholds. Secretome genes were used as the true positive set and non-secretome genes as the true negative set. The AUC (area under the curve) of SECReTE10 was the highest. B. SECReTE is highly abundant in the mRNAs of human secretome proteins. SECReTE10 abundance was calculated for each codon position separately. SECReTE abundance in human mSMPs is most significant in the second position of the codon, but highly significant differences were also detected in the third position. *p≤ 3.73E-68, Chi-square test C. SECReTE is highly abundant in mRNAs coding for soluble secretome proteins in humans. SECReTE10 presence was examined separately for TMD-containing proteins and soluble secreted proteins. A higher fraction of mRNAs coding for soluble secreted proteins (Secretome without TMD; cyan) contains SECReTE in comparison to non-secretome transcripts without a TMD (light gray). The fraction of soluble secreted proteins having SECReTE in the third position is larger than that of TMD-containing non-secretome proteins (NNY) and is significant. n represent the number of genes in each group. *p ≤ 3.49E-12, Chi-square test. (D) SECReTE10 abundance in different groups of human genes. High SECReTE abundance was observed for other secretome protein groups, except tail-anchored (TA) proteins. Mitochondrial mRNAs (Mito) have low SECReTE abundance. Numbers above bars represent the number of genes in each group. (E) SECReTE10 abundance in B. subtilis. SECReTE10 abundance was scored and was observed to be higher in mRNA coding for genes encoding secretome proteins (i.e. SS&TMD, TMD, and SS) as compared to those encoding non-secretome (Non-Sec) proteins. Numbers under bars represent the number of genes in each group. (F) SECReTE10 abundance in S. pombe SECReTE10 abundance was calculated for each codon position separately for TMD-containing proteins and soluble secreted proteins. A higher fraction of mRNAs coding for soluble secreted proteins (Secretome without TMD; cyan) contains SECReTE in comparison to non-secretome transcripts, either with or without a TMD (Non-secretome with TMD; dark gray, Non-secretome without TMD; light gray). The fraction of soluble secreted proteins having SECReTE in the third position is larger than that of TMD-containing non-secretome proteins (NNY) and is significant. n represent the number of genes in each group. n represent the number of genes in each group. * p≤ 5.63E-3, Chi-square test.