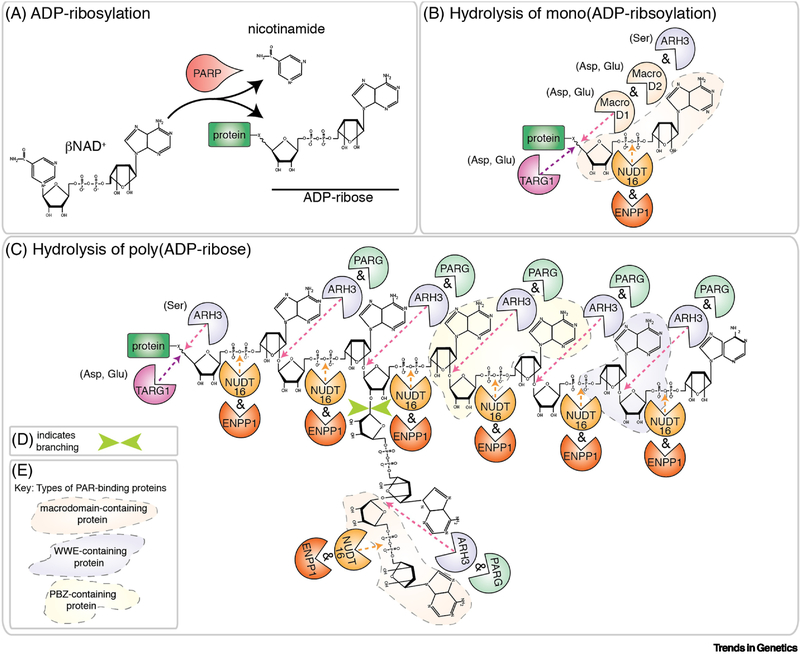
Figure 1: ADP-ribosylation.
A) PARPs catabolize βNAD+ into nicotinamide and ADP-ribose (ADPr), with the resulting ADPr covalently linked onto an acceptor protein. The addition of a single ADPr subunit is called mono(ADP-ribosylation) (MARylation). Amino acids targeted are Arg, Asp, Cys, Glu, Lys, Ser and Tyr [2, 7]. B) Removal of a single ADPr subunit from acidic residues (Asp and Glu) is mediated by MacroD1, MacroD2 and TARG1 [12–14]. ARH3 removes the ADPr from Ser residues [10]. NUDT16 and ENPP1 cleave the 2 phosphate groups in ADPr to generate a phosphoribosylated protein [15–17]. C-D) Poly(ADP-ribosylation) (PARylation) is the sequential addition of ADPr and generates linear and sometimes branched (D, green arrows) polymers of poly(ADP-ribose) (PAR). PARylation is reversed by PARG-mediated hydrolysis of the ribose-ribose bond. ARH3 also cleaves the ribose-ribose bond but unlike PARG, ARH3 can also remove the protein proximal ADPr from Ser residues in the target protein [8–11]. TARG1 removes the entire PAR chain by cleavage of the protein proximal ADPr [14]. The 2 phosphate groups in ADPr are hydrolyzed by NUDT16 and ENPP1 [15–17]. E) Conserved protein domains recognize and bind specific sites of PAR polymers (reviewed in [115]). These domains include the PBZ domain, the WWE domain, the macrodomain and the PAR-binding motif. The physical contacts that are made with the ADPr moiety are known for the PBZ domain, the WWE domain and the macrodomain. Those regions of the PAR chain are shaded in B and C according to the key in E. It is currently unknown what the PBM interacts with in PAR.