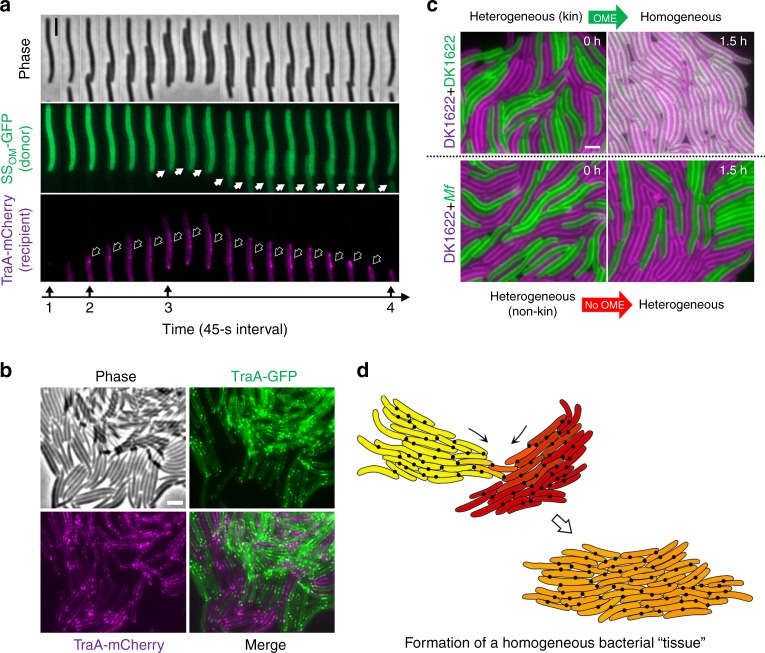
Fig. 5.
TraA/B adhesins transition bacterial kin cells into a tissue. a Representative time series showing TraA recognition followed by the transfer of SSOM-GFP between cells. Marked in the timeline: 1, before contact no foci are detected; 2, upon contact the TraA foci form (hollow arrows); 3, cargo transfer is detected (solid arrows); 4, cell separation and dissolution of TraA foci. b TraA junctions assemble bacteria into a tissue-like structure. TraA-GFP cells were co-incubated with TraA-mCherry cells at a high density (a sub-view of images shown in Supplementary Fig. 11). c TraA recognition governs the bidirectional transfer between cells harboring SSOM-GFP or SSOM-mCherry cellular cargo. Compatible TraA receptors allow exchange after the indicated incubation time (top panels), whereas incompatible receptors block exchange (bottom panels). traA alleles shown at the left. See Supplementary Fig. 12 for a more detailed analysis. Strain details are in Supplementary Table 1. Scale bar = 1 µm. d A schematic illustration of two kin populations that are phenotypically divergent (labeled yellow and red) that recognize and merge into a homogeneous tissue-like population through OME (orange). The extracellular matrix (polysaccharides and proteins, not shown here) also contribute to tissue formation. Black dots represent TraA/B foci located at cell–cell contacts