Fig. 5.
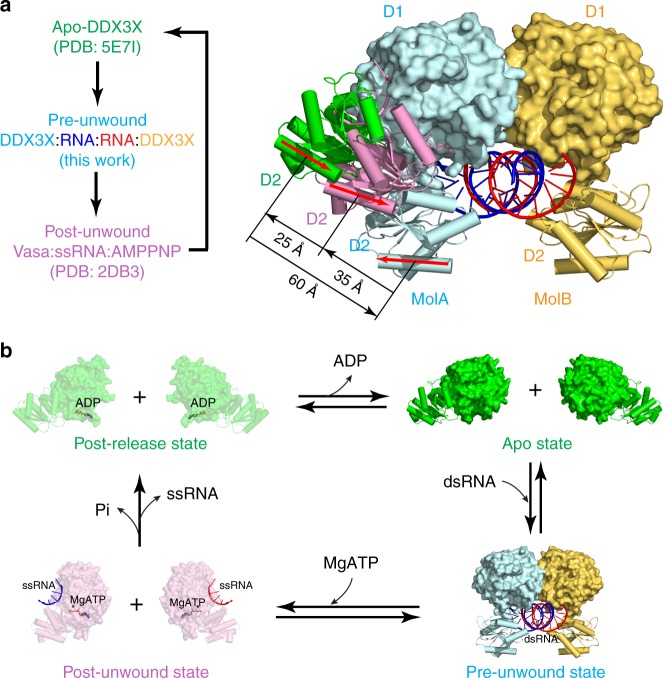
Unwinding of double-stranded RNA (dsRNA) by DEAD-box helicases. a Conformational difference between the apo-DDX3X (in green, PDB: 5E7I), DDX3X:dsRNA:DDX3X (in cyan and orange, this work), and Vasa:MgAMPPNP:ssRNA (in pink, PDB: 2DB3) structures. The structures are aligned based on Cα positions in D1. The D1 of the DDX3X:dsRNA:DDX3X structure is illustrated as a molecular surface, and those of other structures are not shown for clarity. The D2 of all three structures are shown as cartoon models (helices as cylinders, strands as arrows, and loops as tubes). The orientation of D2 relative to the fixed D1 at each state are indicated with a red arrow on the same α-helix in D2. b Represented by the apo (PDB: 5E7I), pre-unwound (this work), post-unwound (PDB: 2DB3), and post-release (PDB: 4PXA) states, the functional cycle explains a four-step mechanism of RNA duplex unwinding by DEAD-box helicases