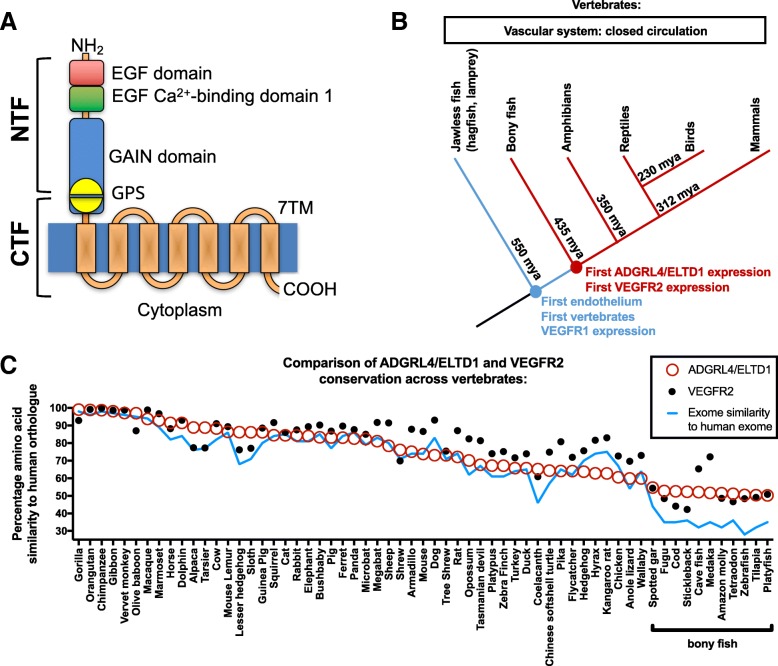
Fig. 1.
ADGRL4/ELTD1 is encoded in the genomes of bony fish and all subsequent vertebrates suggesting that it became established in vertebrates approximately 435 mya at the same time as VEGFR2. a Schematic representation of human ADGRL4/ELTD1’s putative structure showing NTF and CTF division. The NTF comprises an EGF-like domain and a Ca2+ binding EGF-like domain as well as the majority of the GAIN domain and N-terminal portion of the GPS. The CTF comprises the C-terminal portion of the GPS as well as the 7TM domain and a short intracellular ICD. b Cladogram depicting vertebrate evolution and the presence of orthologues to genomic ADGRL4/ELTD1 and the core angiogenetic genes VEGFR1 and VEGFR2. Blue represents the presence of orthologues to genomic VEGFR1 without any VEGFR2 or ADGRL4/ELTD1. Red indicates the presence of orthologues to genomic VEGFR1, VEGFR2 and ADGRL4/ELTD1. Evolutionary divergence time estimates from [25, 26]. Cladogram not drawn to scale. c Comparison of ADGRL4/ELTD1 and VEGFR2 orthologue amino acid conservation across 61 vertebrate species with fully sequenced and annotated genomes reveals high ADGRL4/ELTD1 conservation across all vertebrates with both ADGRL4/ELTD1 and VEGFR2 following a similar trend. Each data point represents the percentage similarity of orthologue to human ADGRL4/ELTD1 (red circle) or VEGFR2 (black dot). The blue line (representing the percentage similarity of each species’ exome contrasted to the human exome) reveals that in bony fish, ADGRL4/ELTD1’s conservation is significantly higher than the exome conservation similarity suggesting that ADGRL4/ELTD1 is part of an early core angiogenic gene group and confirming previous in vivo experiments. (Abbreviations: GAIN = GPCR autoproteolysis inducing domain; GPS = GPCR proteolysis site)