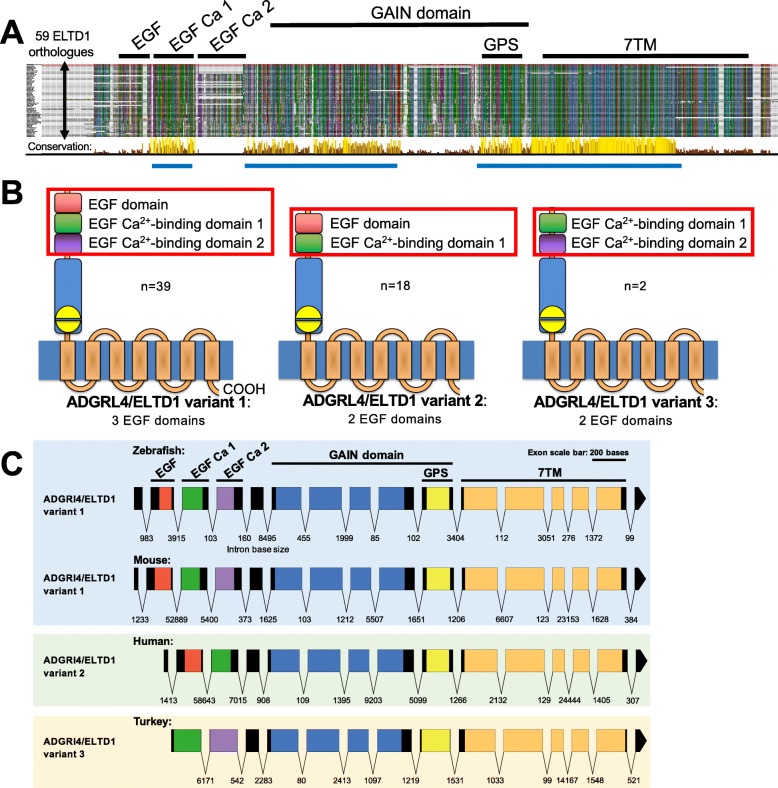
Fig. 2.
ADGRL4/ELTD1 comprises three evolutionary variants. a amino acid conservation across 59 extant vertebrate orthologues reveals 3 evolutionary ADGRL4/ELTD1 variants and three areas of major conservation: i) the EGF Ca binding domain; ii) GAIN domain and GPS motif; and iii) 7TM domain. Conservation scores are presented in the conservation track below the alignments; the highest scores being presented as yellow tall bars and the lowest with no bars. These results highlight ADGRL4/ELTD1’s 2 main evolutionary variants: the oldest (and majority) of ADGRL4/ELTD1 sequences comprise three EGF domains, whilst the more evolutionary recent species encode only 2 EGF domains. ADGRL4/ELTD1’s domains are annotated with black bars. Highly conserved areas are annotated with blue bars. Aligned genomes are listed starting with human (primate) continuing down the evolutionary ladder until platyfish (bony fish). b Graphical summary of ADGRL4/ELTD1’s three evolutionary variants showing the frequency of each variant in the sample of orthologues examined. c Exon map of representative vertebrates encoding ADGRL4/ELTD1 variants 1–3 showing that each EGF and EGF Ca binding domain segregates to its own exon. In vertebrates which only express ADGRL4/ELTD1 variant 2, during evolution the fourth exon was likely deleted from ADGRL4/ELTD1 variant 1 producing ADGRL4/ELTD1 variant 2. In the case of ADGRL4/ELTD1 variant 3, the first two exons were removed. Exons are drawn to scale whilst intron sizes are listed between each exon and are not drawn to scale. Domains are listed and are colour coded