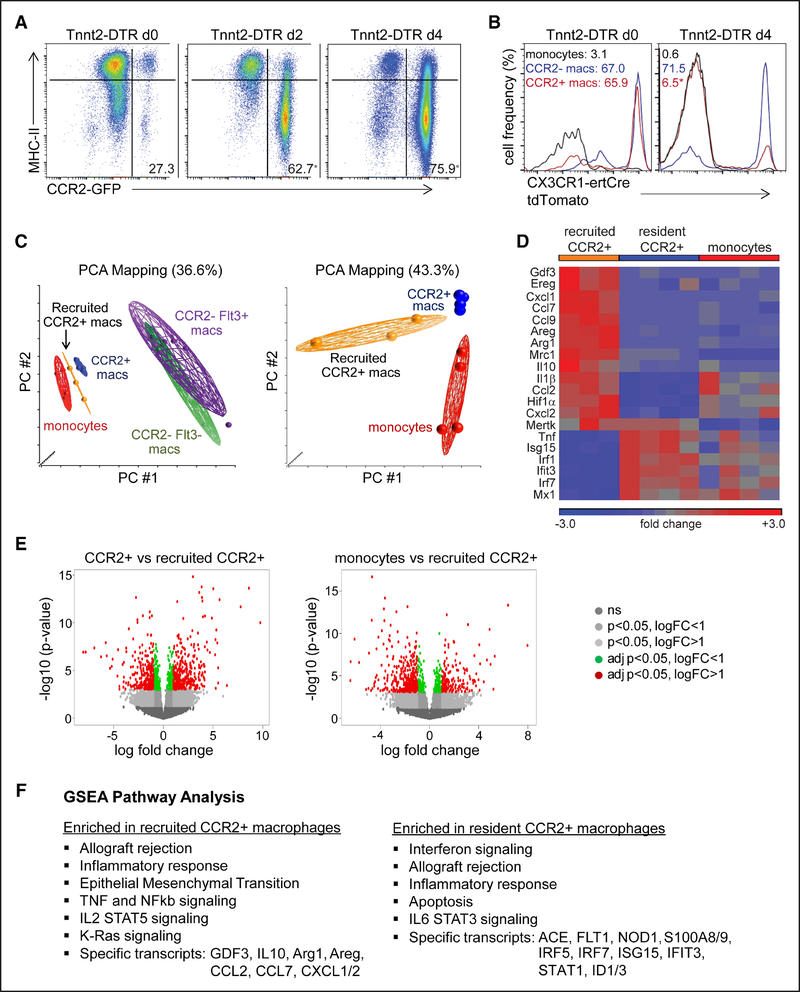
Figure 2. Recruited monocyte-derived macrophages are distinct from tissue-resident subsets.
A and B, Flow cytometry of cardiac monocyte and macrophage subsets in Tnnt2 (troponin T2)-DTR (diphtheria toxin receptor; A) and Tnnt2-DTR CX3CR1ertCre (CX3C chemokine receptor 1) Rosa26tdTomato (B) mice after diphtheria toxin administration and the TAM pulse-chase protocol. Displayed frequencies indicate the percentage of CCR2+ (C-C chemokine receptor 2) monocytes and macrophages (left) and the percentage of tdTomato+ monocytes and macrophages (right). n=4 per experimental group. C, Principal component analysis (PCA) of RNA sequencing data obtained from sorted monocyte and macrophage populations harvested from the hearts of Tnnt2-DTR Flt3-Cre Rosa26tdTomato mice after diphtheria toxin administration. Tissue-resident CCR2− and CCR2+ macrophages were isolated from hearts 36 h after diphtheria toxin (DT) treatment, recruited CCR2+ macrophages were isolated from hearts 4 d after DT treatment. Monocytes were harvested 36 h and 4 d after DT treatment. D, Heat map highlighting genes that were differentially expressed between tissue-resident CCR2+ macrophages, recruited CCR2+ macrophages, and CCR2+Ly6Chigh monocytes. E, Volcano plots showing the number of genes differentially expressed between tissue-resident CCR2+ vs recruited CCR2+ macrophages (left) and monocytes vs recruited CCR2+ macrophages (right). logFC: log based 2 fold change, adj p: adjusted P (false discovery rate analysis). F, Gene set enrichment analysis (GSEA) pathway analysis showing pathways and specific genes enriched in tissue-resident and recruited CCR2+ macrophages. *P <0.05 compared with baseline.