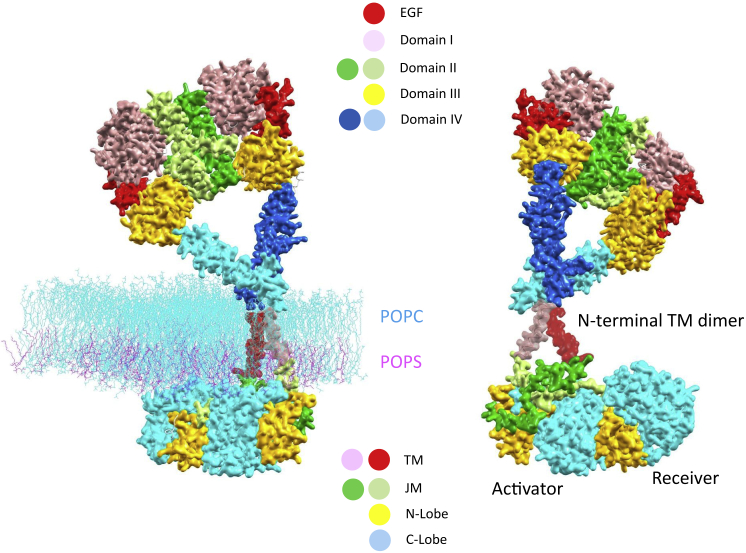
Figure 4.
A near-complete atomic model of the active ligand-bound EGFR dimer in the membrane. Depicted here is a snapshot of the atomic model from MD simulations, which was put together by Shaw and co-workers (7) based on known EGFR segmented structures excluding the disordered part of the C-terminal autophosphorylation CT substrate. On the left, the lipid bilayers included in the simulation (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine in the wire-frame model by the colors cyan and magenta, respectively) specify the position of the membrane with respect to active EGFR. Shown on the right is a view after a 180° rotation along the paper axis, purposely excluding the lipid bilayers to clearly reveal structurally coupled conformations responsible for physiological EGFR activation, including the TM near the N-terminal-cross helical dimer, the antiparallel JM-A helical dimer, and the key interaction of the receiver JM-B with the activator kinase C-lobe. To see this figure in color, go online.