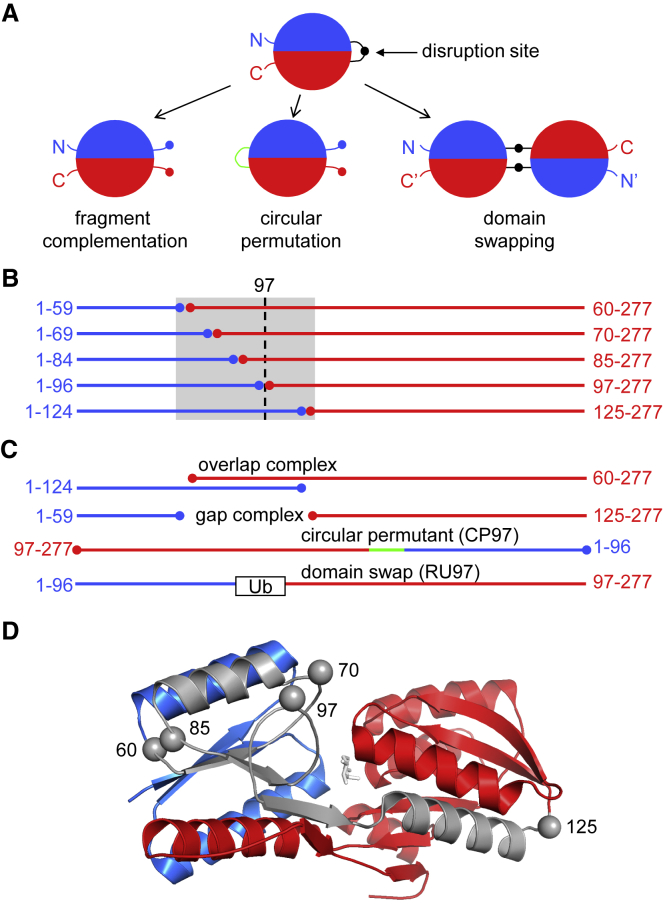
Figure 1.
Fragment complementation, circular permutation, and domain swapping. (A) These three protein engineering methods begin by disrupting a protein’s sequence at a position denoted by the black circle (typically within a surface loop). The linker peptide in the circular permutant and the hinge region in the domain-swapped dimer are colored green and black, respectively. (B) The five N-terminal fragments of RBP (blue) and five C-terminal fragments (red) created for this study are represented by their amino-acid sequences and arranged as pairs of exactly complementary fragments (homo complexes). The gray box demarcates the region duplicated in the overlap complexes and absent in the gap complexes. Position 97 is indicated by the dashed line. (C) The complexes with the longest segments of duplicated sequence (RBP1–124 + RBP60–277) and missing sequence (RBP1–59 + RBP125–277) are shown to illustrate overlap and gap complexes, respectively. Circular permutation (CP97) and Ub insertion (RU97) at position 97 are depicted with linker peptide in green and Ub as a box, respectively, as the lower two sequences. (D) Locations of split sites in RBP (Protein Data Bank [PDB]: 2IOY) are shown as gray spheres. Colors represent the same regions as in (B). Ribose is shown as white sticks.