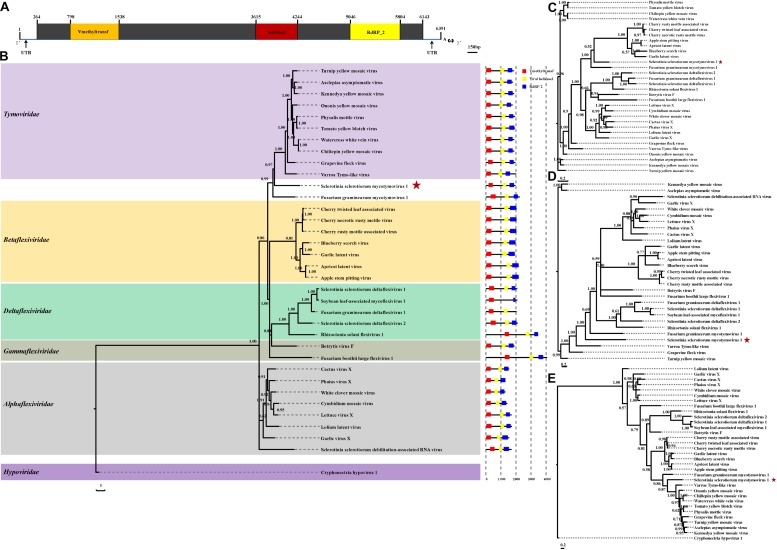
FIGURE 2.
Analyses of identified Sclerotinia sclerotiorum mycotymovirus 1 (SsMTV1/SZ-150) genome. (A) Schematic diagrams of the genetic organization of SsMTV1/SZ-150. The ORF (nt positions 264–6,143) encodes a putative viral protein with 1,958 amino acid residues that contains three conserved domains represented by the box: Vmethyltransf (798–1,538 nt), helicase (3,615–4,244 nt), and RNA-dependent RNA polymerase (RdRP_2, 5,046–5,804 nt). 5′-UTR (nt positions 1–263 nt) and 3′-UTR (nt positions 6,144–6,391 nt) are indicated as lines. The scale bar corresponds to a length of 150 bp. Phylogenetic (Bayesian) trees were constructed from the sequences of three conserved domains of the entire RP domain (B), helicase (C), methyltransferase (D), RdRP (E), and the SsMTV1/SZ-150, with the models of VT+I+G+F, RtREV+I+G+F, LG+I+G+F, and LG+I+G+F, respectively. CHV1 (Cryphonectria hypovirus 1) served as an outgroup in the phylogenetic analysis. The scale bar indicates the number of amino acid substitutions per site; numbers at branch nodes show percentage posterior probabilities. The position of SsMTV1/SZ-150 is shown by red asterisks. The detailed information on the selected viruses in the phylogenetic tree is presented in Table 1.