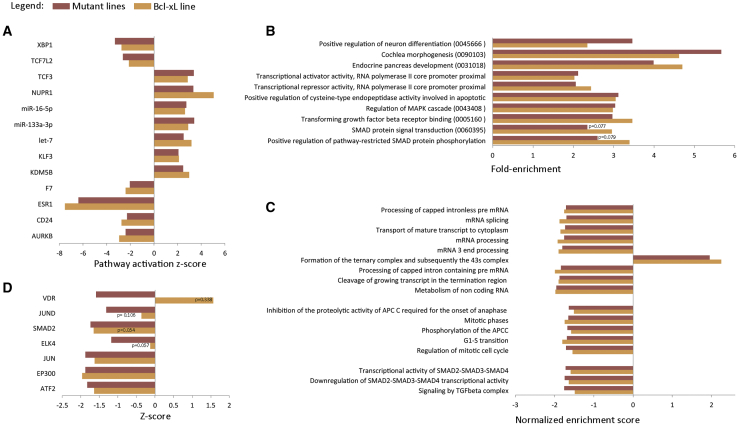
Figure 2.
Pathway Analysis in hESCs with a Gain of 20q11.21 and in VUB03_BcL-xL
(A) Ingenuity Pathway Analysis of the upstream regulators of pathways with an activation score between −2 and 2 and a p value < 0.05 in the wild-type (WT) versus mutant (MT) lines and their behavior in VUB03_BcL-xL.
(B) DAVID Enrichment Analysis of the 1,000 top deregulated genes in the mutant group and VUB03_BcL-xL. In parenthesis are the gene-ontology term numbers.
(C and D) Gene set enrichment analysis using the MSigDB C2 library (C) and Enrichr-based prediction of protein-protein interactions for transcription factors (D). All results are statistically significant with a p value < 0.05 unless the value is given. All the analyses were performed using the differentially expressed genes between WT versus mutant group and WT versus VUB03_BcL-xL with a cutoff value of |log2 fold change| > 1 and FDR < 0.05.