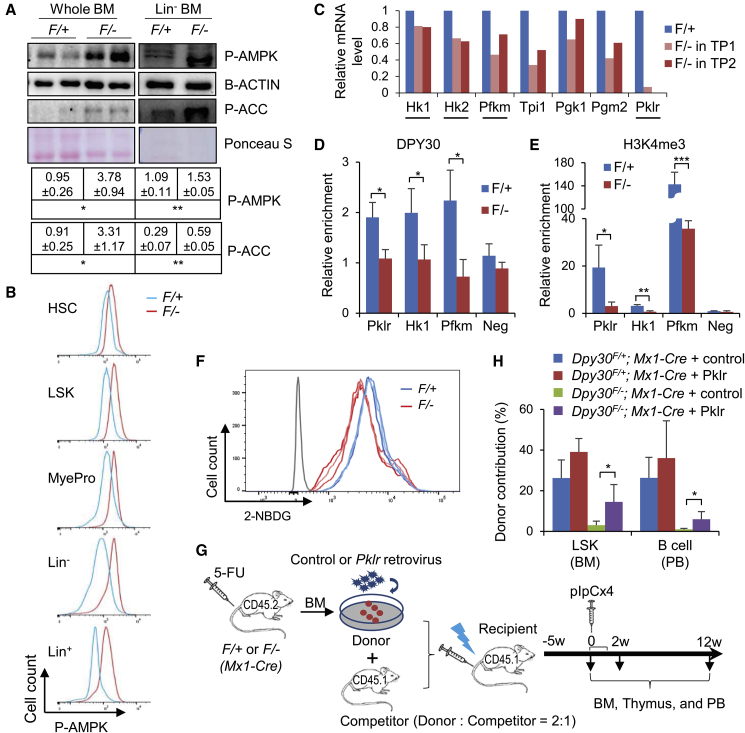
Figure 2.
DPY30 Deficiency Impairs Glucose Metabolism in BM HSPCs
(A) Immunoblotting in whole or Lin− BMs from pIpC-injected Mx1-Cre; Dpy30F/+ and Mx1-Cre; Dpy30F/− mice. Each lane represents an individual pool of cells. Bottom, quantification from three animals each.
(B) Representative fluorescence-activated cell sorting (FACS) analyses for phospho-AMPK (Thr172) in different cell populations in BMs from pIpC-injected Mx1-Cre; Dpy30F/+ and Mx1-Cre; Dpy30F/− mice.
(C) Relative expression of glycolytic genes, as analyzed by RNA-seq in LT-HSCs from two independent BM transplantations from previous work (Yang et al., 2016). Underlined genes are rate-limiting for glycolysis.
(D and E) Chromatin immunoprecipitation for DPY30 (D) and H3K4me3 (E) using Lin− BM cells from pIpC-injected Mx1-Cre; Dpy30F/+ and Mx1-Cre; Dpy30F/− mice. n = 3 mice each.
(F) Representative histogram showing reduced 2-NBDG uptake in vitro by sorted LSK cells from pIpC-injected Mx1-Cre; Dpy30F/+ and Mx1-Cre; Dpy30F/− mice. The gray line indicates isotype control. Each colored line represents an individual mouse out of three mice each.
(G) Scheme for the rescue assay using BMs from Mx1-Cre; Dpy30F/+ (F/+) and Mx1-Cre; Dpy30F/− (F/−) mice. Both the control and Pklr viral constructs expressed GFP.
(H) Donor contribution to indicated cell populations in the transplant recipients 2 weeks after pIpC-injections. n = 4 mice each.
Data are shown as mean ± SD for (A, D, E, and H). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, by two-tailed Student's t test for (A, D, and E), and by one-factor ANOVA with post hoc t test for (H). See also Figure S3.