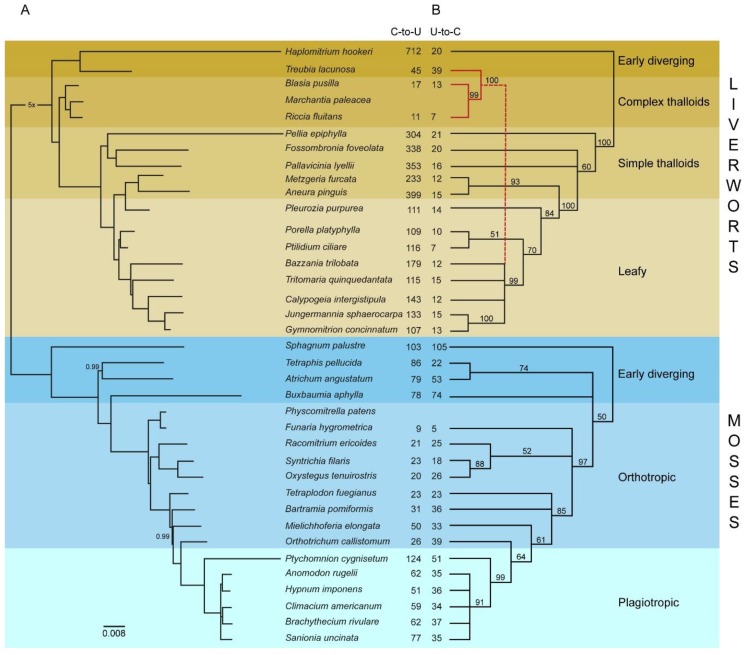
Figure 1.
Phylogenetic relationships of 37 species of liverworts and mosses based on 33 concatenated mitochondrial protein-coding sequences and predicted RNA editing sites. (A) Phylogenetic tree obtained as a result of Bayesian inference analysis of 33 concatenated protein-coding sequences of 37 species. All branches are maximally supported unless otherwise marked. The scale bar indicates the number of substitutions per nucleotide position. (B) Phylogenetic tree obtained as a result of maximum parsimony analysis of binary matrix of predicted C-to-U RNA editing site occurrence within the 33 aforementioned protein-coding sequences of 35 species. The support values are given at the nodes. Two columns in the middle of the graph depict the total number of C-to-U and U-to-C editing sites predicted within the aforementioned species sequences.