Abstract
Sixty strains (n = 60) of Vibrio vulnificus were examined for their multiple antibiotic resistance (MAR) index, plasmid profiles, and DNA polymorphisms. Thirty-seven strains (n = 37) were isolated from cockles (Anadara granosa) in Malaysia, while 23 (n = 23) isolates were isolated from clams (Mercenaria mercenaria) in Qatar. All isolates were resistant to two or more of the antibiotics tested, with the most common resistances were demonstrated towards penicillin (93%), ampicillin (70%), cephalothin (65%), clindamycin (66%), vancomycin (64%), and erythromycin (51%). The antibiotic that experienced the least resistance was kanamycin (6%), and all isolates were susceptible to cefoperazone, streptomycin, and tetracycline. The MAR index for the V. vulnificus isolated from Malaysia and Qatar, possessed similar values which ranged from 0.2 to 0.7, respectively. Plasmid analysis demonstrated that 65% of V. vulnificus strains harbored plasmids, while 35% were not. Nineteen (P1–P19) plasmids profiles were observed. No specific cluster or group was observed although they were isolated from different sample sources and locations by phylogenetic analysis using GelCompar II software at an 80% similarity level. Results demonstrated the high MAR index and genomic heterogeneity of V. vulnificus, which are of great concern to the human health of those who have consumed cockles and clams from the study area.
Keywords: multiple antibiotic resistance (MAR) index, plasmid profiles, DNA polymorphisms, Vibrio vulnificus, genomic heterogeneity
1. Introduction
Vibrio vulnificus is a rod-shaped, Gram-negative halophile and an opportunistic human pathogen. It belongs to the family of Vibrionaceae and is ubiquitous in marine environments. This bacterium has been isolated from water, sediments, fish, and shellfish [1,2]. V. vulnificus can cause diseases to individuals who eat contaminated seafood or have an open wound infected by this bacterium via seawater. Their infection can be fatal or can cause sepsis in susceptible individuals.
In the previous studies, potentially pathogenic Vibrio species such as Vibrio parahaemolyticus, V. cholera, and V. vulnificus were detected in seafood sold in Malaysia, the later was recently found in shrimps, squids, crabs, cockles, and mussels. The results of these studies showed that the most virulent of the non-cholera vibrios—V. vulnificus—has various virulence factors that facilitate the development of clinical disease [3,4]. On the other hand, no incidence of V. vulnificus was reported in Qatar. An investigation is greatly needed because of the current increasing concern that V. vulnificus may represent a clinical problem, especially in communities that consume shellfish such as cockles, oysters, and clams.
Antibiotic resistance is used as an epidemiological tool for foodborne disease control; it also provides antibiotic information, which may help to treat disease due to this bacterium. The emergence of multiple antibiotics resistance (MAR) bacteria may pose a threat to human health [5]. The emergence of MAR bacteria was due to indiscriminate use of antibiotics in clinical medicine, agriculture and aquaculture industries [6]. Vibrio spp. was reported to be greatly susceptible to the majority of clinically used antibiotics [7]. However, based on annual reports, an increasing number of Vibrio spp. have become more resistant toward clinically utilized antibiotics [8]. Antibiotics may contribute to the survival of bacteria strains that may contain resistance (R) plasmids. The transfer of R plasmids from resistant to nonresistant organisms is of great medical significance because it reduces the effective use of antibiotics. A previous study reported that there was a correlation between antibiotic resistance and the presence of the plasmids on Vibrio spp. [9]. Strains of biotype 2 of V. vulnificus possess one or more virulence plasmids [10], ranging between 68 and 70 kb. V. vulnificus strains were also found to carry more than one plasmid with diverse sizes [11].
Molecular approaches are useful to enable us to group bacteria into distinctive groups according to their distinctive features, between two geographical locations. Phenotype-based subtyping, such as antibiotic resistance, and deoxynucleic acid (DNA)-based subtyping, such as plasmid profiles and random amplified polymorphism DNA (RAPD) analysis, allow the bacterial isolates to be differentiated below the species level. Bacterial subtyping would help researchers to detect and track foodborne disease outbreaks, clonal species circulating in different locations, and as tools to track the sources of bacterial contamination in the food system. It also facilitates a better understanding of the ecology of different foodborne pathogens, population genetics, and epidemiology. In the present study, we investigated the antibiotic resistance, plasmid profiles and polymerase chain reaction (PCR)-based analysis of V. vulnificus isolated from cockles and clams from Malaysia and Qatar.
2. Results
2.1. Antibiotic Resistance
In general, a total of 60 V. vulnificus isolates from both countries showed resistance towards antibiotics in the following order (Table 1); penicillin (93%), ampicillin (70%), clindamycin (66%), cephlothin (65%), vancomycin (64%), bacitracin (59%), erythromycin (57%), novobiocin (46%), and kanamycin (6%). Most of the isolates in this study were sensitive to cefoperazone, streptomycin and tetracycline. Table 2 shows the antibiogram of 60 V. vulnificus isolates, 37 isolates from Malaysia (cockles) indicated 14 patterns (A1–A14), with the most frequent patterns being A3 (AmpKfP) and A4 (AmpBDaEKfNvPVa). While 23 V. vulnificus isolates from Qatar (clams) showed four new patterns (pattern A15–A18), with A1 (BDaEKfNvPVa) and A14 (AmpBDaEPVa) being the most frequently observed. By combining the two locations, 18 antibiograms were revealed (A1–A18). In the present study, Malaysia and Qatar were chosen, because to compare if there were any differences in term of multiple resistant index (MAR), plasmid profiles and DNA heterogeneities between two distantly geographical locations of V. vulnificus isolates.
Table 1.
Frequency of antibiotic resistance Vibrio vulnificus isolates from cockle and clam samples.
| Antibiotic Tested | Percentage of Resistance (%) | ||
|---|---|---|---|
| Cockle Samples, Malaysia (n = 37) | Clam Samples, Qatar (n = 23) | Mean Average | |
| Ampicillin | 65 | 74 | 70 |
| Bacitracin | 59 | 52 | 56 |
| Cefoperazone | 0 | 0 | 0 |
| Cephalothin | 73 | 57 | 65 |
| Clindamycin | 70 | 61 | 66 |
| Erythromycin | 57 | 45 | 51 |
| Kanamycin | 0 | 13 | 6 |
| Novobiocin | 57 | 35 | 46 |
| Penicillin | 95 | 91 | 93 |
| Streptomycin | 0 | 0 | 0 |
| Tetracycline | 0 | 0 | 0 |
| Vancomycin | 62 | 65 | 64 |
Table 2.
Multiple antibiotic resistance (MAR), antibiotic resistance, plasmid profiling, and random amplified polymorphic DNA (RAPD) analysis of Vibrio vulnificus isolated from cockles (Anadara granosa) from Malaysia and clams (Mercenaria mercenaria) from Qatar.
| Strain No. a | Locations | MAR Index | Antibiotic Patterns b | Plasmid Profiles c | RAPD | Strain Types d | ||
|---|---|---|---|---|---|---|---|---|
| RAPD 11 | RAPD 15 | |||||||
| 1 | Malaysia | 0.53 | BDaEKfNvPVa | A1 | P1 | C1 | F1 | 1 |
| 2 | Malaysia | 0.53 | BDaEKfNvPVa | A1 | P2 | C1 | F1 | 2 |
| 3 | Malaysia | 0.5 | AmpDaEKfNvP | A2 | P2 | C2 | F2 | 3 |
| 4 | Malaysia | 0.53 | BDaEKfNvPVa | A1 | P2 | C3 | F3 | 4 |
| 5 | Malaysia | 0.25 | AmpKfP | A3 | P2 | C4 | F4 | 5 |
| 6 | Malaysia | 0.66 | AmpBDaEKfNvPVa | A4 | P3 | C4 | F5 | 6 |
| 7 | Malaysia | 0.5 | BDaKfNvPVa | A5 | P4 | C5 | F6 | 7 |
| 8 | Malaysia | 0.5 | BDaKfNvPVa | A5 | - | C6 | F7 | 8 |
| 9 | Malaysia | 0.53 | AmpBDaKfNvPVa | A6 | - | C6 | F8 | 9 |
| 10 | Malaysia | 0.25 | AmpKfP | A3 | P5 | C7 | F9 | 10 |
| 11 | Malaysia | 0.35 | AmpKfP | A3 | P3 | C8 | F10 | 11 |
| 12 | Malaysia | 0.16 | PVa | A7 | P1 | C8 | F11 | 12 |
| 13 | Malaysia | 0.66 | AmpBDaEKfNvPVa | A4 | P6 | C9 | F12 | 13 |
| 14 | Malaysia | 0.66 | AmpBDaEKfNvPVa | A4 | - | C10 | F13 | 14 |
| 15 | Malaysia | 0.5 | AmpDaEKfNvP | A2 | - | C11 | F14 | 15 |
| 16 | Malaysia | 0.25 | AmpKfP | A3 | - | C12 | F15 | 16 |
| 17 | Malaysia | 0.25 | AmpKfP | A3 | - | C13 | F16 | 17 |
| 18 | Malaysia | 0.58 | AmpBDaKfNvPVa | A6 | P7 | C14 | F17 | 18 |
| 19 | Malaysia | 0.66 | AmpBDaEKfNvPVa | A4 | P8 | C15 | F18 | 19 |
| 20 | Malaysia | 0.25 | KfPVa | A8 | P9 | C15 | F19 | 20 |
| 21 | Malaysia | 0.58 | AmpBDaKfNvPVa | A6 | P10 | C16 | F20 | 21 |
| 22 | Malaysia | 0.66 | AmpBDaEKfNvPVa | A4 | - | C16 | F20 | 22 |
| 23 | Malaysia | 0.66 | AmpBDaEKfNvPVa | A4 | P2 | C16 | F21 | 23 |
| 24 | Malaysia | 0.25 | AmpPVa | A12 | - | C16 | F21 | 24 |
| 25 | Malaysia | 0.25 | AmKfP | A3 | P8 | C17 | F22 | 25 |
| 26 | Malaysia | 0.33 | BDaEP | A9 | P2 | C18 | F23 | 26 |
| 27 | Malaysia | 0.5 | BDaENvPVa | A10 | P3 | C19 | F24 | 27 |
| 28 | Malaysia | 0.33 | AmpBDaE | A11 | - | UT | F25 | 28 |
| 29 | Malaysia | 0.66 | AmpBDaEKfNvPVa | A4 | P3 | C20 | F25 | 29 |
| 30 | Malaysia | 0.33 | BDaEP | A9 | P3 | UT | F26 | 30 |
| 31 | Malaysia | 0.25 | AmpPVa | A12 | P1 | C21 | F27 | 31 |
| 32 | Malaysia | 0.5 | AmpDaEKfNvP | A2 | P11 | C22 | F28 | 32 |
| 33 | Malaysia | 0.5 | BDaENvPVa | A10 | P3 | C23 | F29 | 33 |
| 34 | Malaysia | 0.16 | KfVa | A13 | P3 | C24 | F29 | 34 |
| 35 | Malaysia | 0.5 | AmpBDaEPVa | A14 | - | C25 | F30 | 35 |
| 36 | Malaysia | 0.33 | BDaEP | A9 | - | C26 | F31 | 36 |
| 37 | Malaysia | 0.5 | AmpDaEKfNvP | A2 | P7 | C27 | F32 | 37 |
| 38 | Qatar | 0.66 | AmpBDaEKKfNvP | A15 | P12 | C28 | F33 | 38 |
| 39 | Qatar | 0.41 | AmpKNvPVa | A16 | P13 | C28 | F34 | 39 |
| 40 | Qatar | 0.5 | AmpDaEKfNvP | A2 | - | C29 | F35 | 40 |
| 41 | Qatar | 0.41 | AmpDaKPVa | A17 | - | C30 | F36 | 41 |
| 42 | Qatar | 0.25 | AmpPVa | A12 | P14 | C31 | F37 | 42 |
| 43 | Qatar | 0.5 | AmpBDaEPVa | A14 | - | C32 | F38 | 43 |
| 44 | Qatar | 0.25 | AmpPVa | A12 | P3 | C34 | F40 | 44 |
| 45 | Qatar | 0.5 | AmpBDaEPVa | A14 | P6 | C35 | F41 | 45 |
| 46 | Qatar | 0.25 | AmpKfP | A3 | - | C16 | F21 | 46 |
| 47 | Qatar | 0.58 | AmpBDaKfNvPVa | A6 | - | C36 | F42 | 47 |
| 48 | Qatar | 0.41 | AmpBDaEP | A18 | P15 | C36 | F43 | 48 |
| 49 | Qatar | 0.25 | AmpKfP | A3 | - | C37 | F43 | 49 |
| 50 | Qatar | 0.5 | AmpBDaEPVa | A14 | - | C38 | F44 | 50 |
| 51 | Qatar | 0.58 | BDaEKfNvPVa | A1 | P16 | C39 | F45 | 51 |
| 52 | Qatar | 0.58 | BDaEKfNvPVa | A1 | P17 | C40 | F46 | 52 |
| 53 | Qatar | 0.58 | BDaEKfNvPVa | A1 | P18 | C41 | F47 | 53 |
| 54 | Qatar | 0.5 | AmpBDaEPVa | A14 | P16 | C42 | F48 | 54 |
| 55 | Qatar | 0.5 | AmpBDaEPVa | A14 | - | C43 | UT | 55 |
| 56 | Qatar | 0.25 | AmpKfP | A3 | P17 | C44 | F49 | 56 |
| 57 | Qatar | 0.25 | AmpKfP | A3 | P18 | C45 | F50 | 57 |
| 58 | Qatar | 0.16 | KfVa | A13 | - | ND | F50 | 58 |
| 59 | Qatar | 0.16 | KfVa | A13 | P19 | C46 | F50 | 59 |
| 60 | Qatar | 0.58 | BDaEKfNvPVa | A1 | P15 | C47 | F50 | 60 |
aV. vulnificus isolates 1–37 isolated from Malaysia, 38–60 isolates from Qatar. b Tested for Ampicillin (Amp), Bacitracin (B), Cephalothin (Kf), Clindamycin (Da), Erythromycin (E), Kanamycin(K), Novobiocin (Nv), Penicillin (P), Streptomycin (S), and Vancomycin (Va). c Tested for plasmid profiles. d Combination between antibiotic resistance, plasmid profiling, and random amplified polymorphism DNA (RAPD) fingerprinting. A1–A18, antibiotic patterns, and P1–P21, plasmid patterns. MAR—multiple antibiotics resistance; ND—not determined; and UT—untypable.
2.2. Multiple Antibiotic Resistance (MAR) Index
V. vulnificus isolates from Malaysia and Qatar had different multiple antibiotic resistance (MAR) indexes, ranging from 0.2 to 0.7, respectively. Coincidently, both V. vulnificus isolates from Malaysia and Qatar had similar MAR values, ranging from 0.2 to 0.7, respectively (Table 3). Table 3 details the percentage of occurrence of each MAR index of V. vulnificus isolates from cockle samples from Malaysia and clam samples from Qatar. Although they have a similar MAR index, the percentage of occurrence in both places was different. A total of 15 V. vulnificus isolates (25%) were resistant to three antibiotics, 14 (23%) to six antibiotics, 12 (20%) to seven antibiotics, eight (13%) to eight, five (8%) to two antibiotics, and three (5%) to both four and five antibiotics, respectively (Table 2).
Table 3.
Multiple antibiotic resistance (MAR) index of Vibrio vulnificus isolates (n = 60) from Malaysia (cockles) and Qatar (clams).
| MAR Index | Percentage of Occurrence (%) | |
|---|---|---|
| Isolates from Malaysia (Cockles) (n = 37) | Isolates from Qatar (Clams) (n = 23) | |
| 0.1 | - | - |
| 0.2 | 22 | 32 |
| 0.3 | 16 | - |
| 0.4 | 3 | 13 |
| 0.5 | 35 | 30 |
| 0.6 | 5 | 17 |
| 0.7 | 19 | 4 |
| 0.8 | - | - |
| 0.9 | - | - |
| 1.0 | - | - |
2.3. Plasmid Profiles
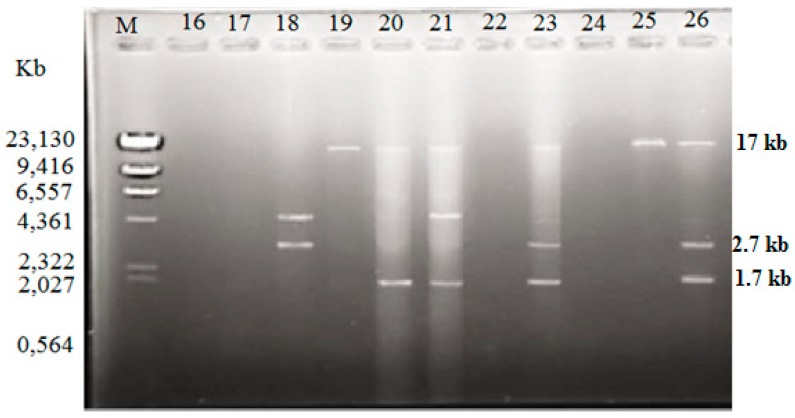
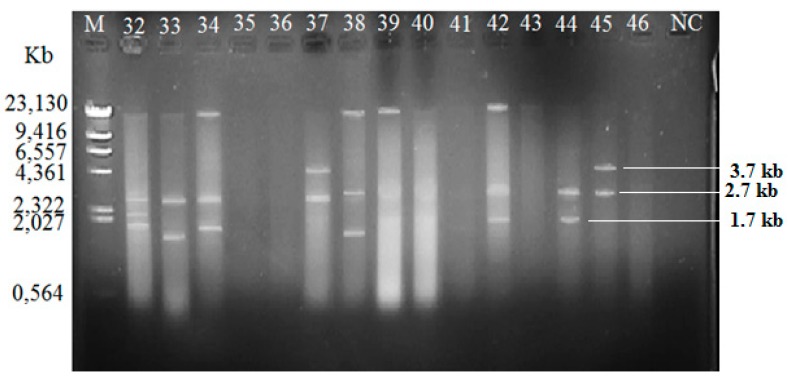
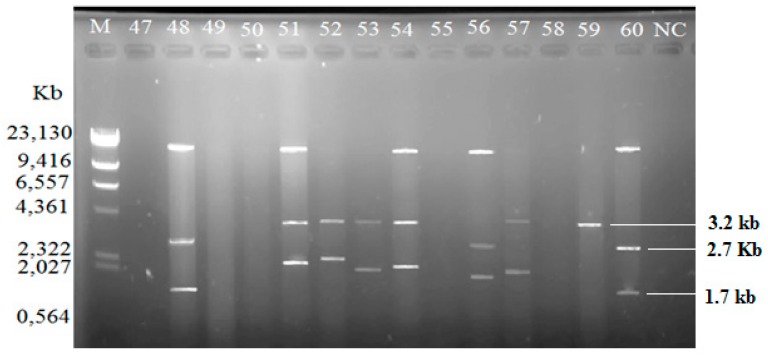
Among the isolated V. vulnificus (n = 60) from Malaysia and Qatar, only 40 (67%) of the strains harbored plasmids, while the other 20 V. vulnificus isolates (33%) did not contain any plasmid (Table 4). Since the Lambda DNA-HindIII DNA ladder is the linear DNA, the determination of plasmids’ molecular weight was based on the plasmid profiling. Plasmid profiles from cockle samples from Malaysia were denoted as P1–P11, while, P3, P6, and P12–P19 represented plasmid profiles from clam samples from Qatar (Table 2). Overall, 19 different plasmid profiles were observed as indicated in Supplementary Materials, Figures S1–S5 and Tables S1–S2. Figure 1, Figure 2 and Figure 3 gives examples of plasmid profiles for several strains of V. vulnificus.
Table 4.
Antibiotic resistance and plasmid occurrence of Vibrio vulnificus isolates from cockle and clam samples purchased from wet markets in Malaysia and Qatar, respectively.
| Antibiotic Tested | Mean Averages (%) Isolates Resistant within Two Sample Sources | No. of V. vulnificus Isolates Resistant | |
|---|---|---|---|
| With Plasmid (40) | Without Plasmid (20) | ||
| Ampicillin | 70 | 36 (90%) | 15 (75%) |
| Bacitracin | 56 | 23 (58%) | 11 (55%) |
| Cefoperazone | 0 | 0 | 0 |
| Cephalothin | 65 | 28 (70%) | 12 (60%) |
| Clindamycin | 66 | 27 (68%) | 13 (65%) |
| Erythromycin | 51 | 22 (55%) | 9 (45%) |
| Kanamycin | 6 | 2 (5%) | 1 (5%) |
| Novobiocin | 46 | 23 (58%) | 6 (30%) |
| Penicillin | 93 | 39 (98%) | 17 (85%) |
| Streptomycin | 0 | 0 | 0 |
| Tetracycline | 0 | 0 | 0 |
| Vancomycin | 64 | 27 (68%) | 11 (55%) |
Figure 1.
Example of plasmid profiles of Vibrio vulnificus isolated from Malaysia (strain 16–26) using a PureYield™ Plasmid Miniprep System on 1% (w/v) agarose gel. Lane M: Lambda DNA-HindIII DNA ladder. Lane 16–26: V. vulnificus strain 16–26.
Figure 2.
Example of plasmid profiles of Vibrio vulnificus isolated from Qatar (strain 32–37, Malaysia; strain 38–46, Qatar) using a PureYield™ Plasmid Miniprep System on 1% (w/v) agarose gel. Lane M: Lambda DNA-HindIII Digest DNA ladder. Lane 47–60: V. vulnificus strain 47–60; NC: negative control.
Figure 3.
Example of plasmid profiles of Vibrio vulnificus isolated from Qatar (strain 47–60, Qatar) using a PureYield™ Plasmid Miniprep System on 1% (w/v) agarose gel. Lane M: Lambda DNA-HindIII Digest DNA ladder. Lane 47–60: V. vulnificus strain 47–60; NC: negative control.
2.4. RAPD-PCR
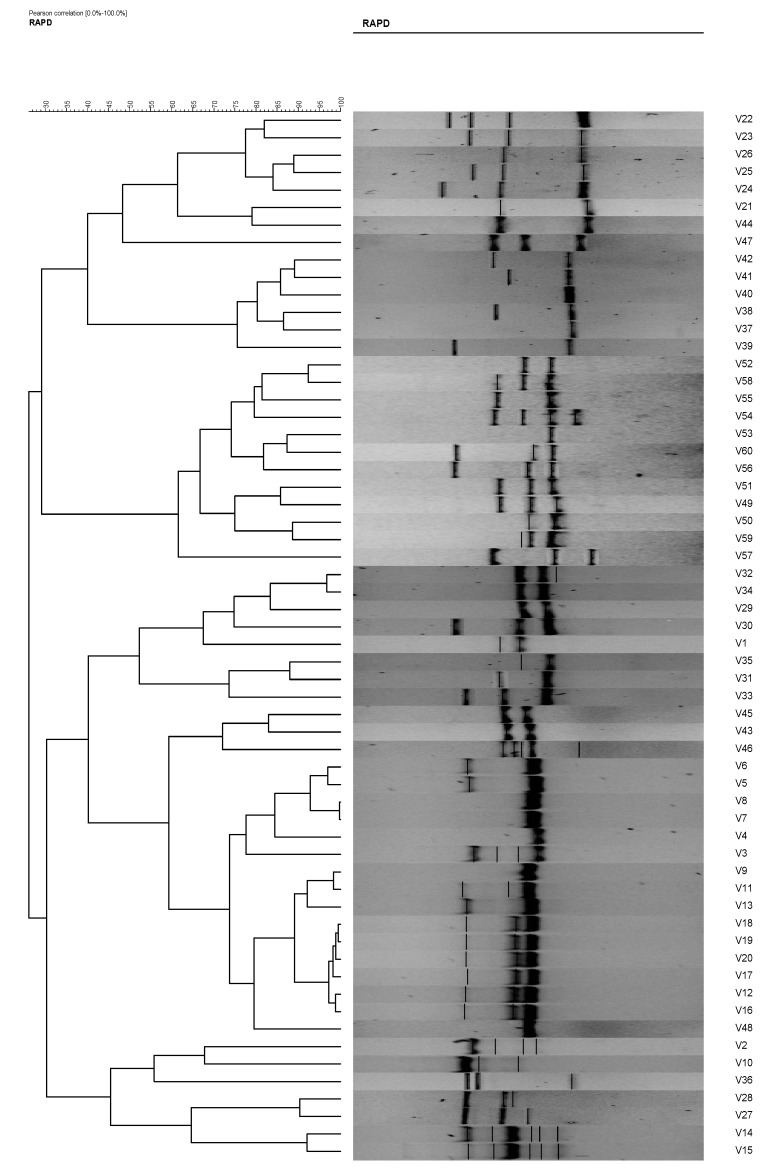
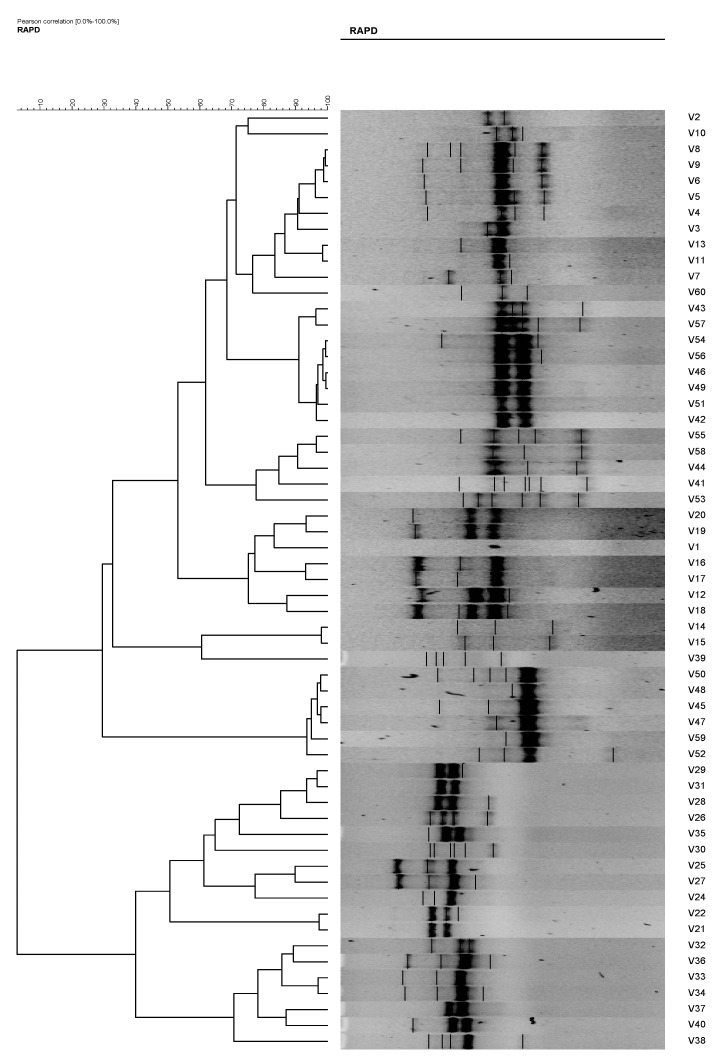
In RAPD analysis, using primer RAPD 11 and RAPD 15, the 60 V. vulnificus isolates generated 47 and 50 RAPD patterns, respectively. Using primer RAPD 11, 27 RAPD patterns were produced by V. vulnificus isolates from Malaysia and are denoted as C1 to C27. From Qatar, 20 RAPD patterns, denoted as C28–C47, were observed (Table 2). Analysis using gel compare II software differentiated the V. vulnificus into 15 clusters and 16 isolates at an 80% similarity level (Figure 4). Primer RAPD15 produced 32 RAPD patterns from V. vulnificus isolates from Malaysia, while 18 patterns were produced by V. vulnificus isolates from Qatar. Using gel compare II analysis, six clusters and three single isolates at the same level were analyzed as shown in Figure 5.
Figure 4.
Phylogenetic tree of Vibrio vulnificus isolates (1–60) from RAPD analysis using primer RAPD11, which is able to differentiate the V. vulnificus into 15 clusters and 16 isolates at an 80% similarity level.
Figure 5.
Phylogenetic tree of Vibrio vulnificus isolates (1–60) from RAPD analysis using primer RAPD15, which is able to differentiate into six clusters and three single isolates at an 80% similarity level.
3. Discussion
The main objective of this study is to determine the antibiotic resistance, plasmid profile, and RAPD analysis of V. vulnificus isolated from cockles (Anadara granosa, Malaysia) and clams (Mercenaria mercenaria, Qatar). As indicated in Table 1, all isolates were tested against several antibiotics and the highest antibiotic resistance was observed towards penicillin (93%), followed by ampicillin (70%), clindamycin (66%), cephalotin (65%), vancomycin (64%), bacitracin (56%), erythromycin (51%), novobiocin (46%), and kanamycin (6%). None of these isolates were found to be resistant to cefoperazone, streptomycin, or tetracycline (Table 2). The isolates from cockle samples from Malaysia demonstrated marginally more resistance to many antibiotics compared to clam samples from Qatar, except for ampicillin and vancomycin. Thus, differences that were found in the antibiotic resistance may depend on the sample source.
The highest percentage of resistance towards penicillin (93%) was observed in the present study. These findings were consistent with the previous work, where the penicillin-resistant vibrio has been reported to be 100% resistance towards penicillin in India leading to concern regarding drug-resistant microbial diseases in aquaculture [6]. Two separate studies by Radu et al. [12,13] found that V. vulnificus is highly resistant to both bacitracin and penicillin in Malaysia, which was in agreement regarding penicillin but in contrast with what was previously known regarding bacitracin. In this study, only 56% of V. vulnificus isolates showed resistance towards bacitracin. Furthermore, ~70% of the isolates were resistant to ampicillin, the second highest resistance level after penicillin. Ampicillin was detected as having a high resistance against V. vulnificus isolated from the Arabian Gulf compared to the other antibiotics tested [11], which is in agreement with this study. The high susceptibility of all isolates against tetracycline and streptomycin in the present study indicated that the V. vulnificus was sensitive to those antibiotics compared to other Vibrio species. Son et al. [13] and Okoh et al. [14] reported some of their vibrio isolates were resistant toward streptomycin and tetracycline.
In this study, the V. vulnificus isolates showed high incidences of antibiotic resistance against more than two or more antibiotics. The multiple antibiotic resistance (MAR) index from cockle samples from Malaysia and clam samples from Qatar showed different MAR index values in the range of 0.2 to 0.7 (Table 3). Most of the isolates proved to be resistant to multiple antibiotics (MAR). Fifteen isolates (25%) were resistant to three antibiotics, 14 (23%) to six antibiotics, 12 (20%) to seven antibiotics, eight (13%) to eight antibiotics, five (8%) to two antibiotics, and three (5%) to both four and five antibiotics, respectively. These findings were similar to those reported by Roig and Amaro, Baker-Austin et al., Tunung et al., and Lee et al. [10,15,16,17], in which it was observed that V. vulnificus isolates were resistant to two or more antibiotics with a high MAR index. A MAR index value higher than 0.2 is said to have originated from high-risk sources of antibiotic contamination where antibiotics are often used, such as from human, commercial poultry farms, swine and dairy cattle [17,18]. The occurrence of antibiotic-resistant V. vulnificus in seafood represents a potential hazard to human health, especially to people who consume seafood that has been improperly prepared.
As indicated in Table 3, comparing between Malaysia and Qatar, although the MAR index from both places was similar (0.2–0.7), the occurrence percentages were different. For example, more V. vulnificus isolates from cockles samples from Malaysia had a MAR index value of 0.7 (19%) compared to clams samples from Qatar (4%). The MAR value index value of 0.7 for both exhibited resistances to eight of the antibiotics tested (AmpBDaEKfNvPVa); however, the number of isolates from cockle samples from Malaysia was almost five times higher compared to clam samples from Qatar. It has been suggested that a high number of isolates from cockle samples from Malaysia may come from the continued agricultural use of medicated feeds in animal husbandry which disseminate the virulent and resistant bacterial pathogens through the feces, resulting in dispersal into the environment. It is possible that the plasmid exchanged between bacteria in aquatic systems would also contribute to the high frequency of MAR incidences [11,14].
Plasmid is one of the most important mediators that facilitate the fast spread of antibiotic resistance among bacteria [18]. The transferal of genetic elements of antibiotic resistance to other bacteria can cause illness in humans [19]. When the resistance of isolates carrying plasmids was compared with that of isolates without plasmids, the results were almost similar for resistance towards bacitracin, clindamycin, erythromycin, and kanamycin. However, more isolates containing plasmids were resistant to ampicillin, cephalotin, novobiocin, penicillin, and vancomycin. As indicated in Table 4, 67% (40/60) of V. vulnificus isolates harbored plasmids, while 23% (20) of isolates did not. Approximately 26 of 37 (70%) of isolates from cockle samples and 14 from 23 (60%) isolates from clam samples harbored plasmid DNAs. This finding suggested that the resistance of V. vulnificus isolates was encoded on plasmids for ampicillin, cephalotin, novobiocin, penicillin, and vancomycin, or on chromosomes for bacitracin, clindamycin, erythromycin, and kanamycin. Aoki et al. [20] reported that the antibiotic resistance of V. anguillarum occurred in R plasmid and genome DNA. Furthermore, the antibiotic resistance of nalidixic acid and furazolidone were not transferred to Escherichia coli, indicating that they were present in genome DNA. However, for chloramphenicol, sulfonamides, streptomycin, ampicillin, and trimethoprim, the transferable R plasmids were carried by the strains. However, no conclusion can be drawn since the conjugation analysis has not been conducted.
The plasmid analysis of all the isolates (Table 2 and Table 4), provide a general picture of plasmids in V. vulnificus isolates. Of the 60 strains analyzed, 40 (67%) of the V. vulnificus strains harbored plasmids and 19 different plasmid profiles were observed. The most frequent plasmid profiles were P3 and P2. The multiple plasmids harbored in V. vulnificus isolates are in agreement with ElHadi [11] and [12], who reported the occurrence of multiple plasmids in V. vulnificus isolates. A total of 19 plasmid profiles were shown to be heterogeneous, suggesting it is useful as a tool for categorizing typing V. vulnificus isolates. The variations in the plasmid size of V. vulnificus strains found in this study support the findings of Zhang et al. and Zhang et al. [21,22] who observed multiple plasmids in Vibrio with variations in size. In the present study, plasmid profiles from cockle samples showed 11 profiles (P1–P11), while plasmid profiles from clam samples were mostly P12 to P19. There were two plasmid profiles from Qatar that were similar to Malaysian plasmid profiles, P3 and P6, which suggested they may have similar molecular weights but may be different in terms of their sequences.
The total number of plasmids in any given bacterial population can affect the results of the isolate analysis. For example, isolates 10 and 11 and isolates 51 and 52. These isolates possessed similar antibiotic resistances; however, their plasmid profiles differed. Without plasmid profiles, we may have believed that the V. vulvificus isolates with similar antibiotic resistances originated from the same ancestral isolates. Eighteen (18) antibiogram and 19 plasmid profiles were revealed in the present study, and by the combination of both methods, 43 strain types were observed. However, when these isolates were analyzed using RAPD analysis, the degree of differentiation among the strains increased, resulting in 60 strain types. The results were as expected and in agreement with [4,9,23,24] who reported the used of the RAPD technique in strains differentiation of Escherichia coli, Bacillus cereus, and Vibrio parahaemolyticus, respectively. RADP analysis showed 47 and 50 RAPD profiles using primer RAPD11 and primer RAPD 15, respectively (Table 3, Figure 4 and Figure 5). It was also found that the RAPD profiles from cockles from Malaysia and clams from Qatar were different, but did not show any specific cluster that differentiated between the two distant geographical locations. Two isolates (strain 28 and 30) were untypable using primer RAPD11, and a single isolate was untypable (Isolate 50) using RAPD15 primer, respectively. These results were probably due to the loss of a specific site in their genome DNA.
4. Materials and Methods
4.1. Vibrio vulnificus
A total of 60 V. vulnificus strains were stored in the Laboratory of Food Sciences, Universiti Kebangsaan Malaysia, Selangor. These strains were previously isolated from two distant countries, Malaysia and Qatar. Thirty-seven were from Malaysia, namely, V. vulnificus strains 1–37, while strains 38–60 were from Qatar (23 strains). All strains were isolated between July 2013 and February 2014.
4.2. Antibiotic Resistance
Antibiotic susceptibility tests were performed by the disc diffusion method on Muller Hinton agar (MHA) (Oxoid, UK) as described by Bauer et al. [25]; antibiotic discs are listed in Table 1. A single colony was cultured in 10 mL of Alkaline Peptone Water (APW) (Oxoid) and incubated overnight at 37 °C. The solution was evenly distributed over the MHA using a sterile cotton bud and was allowed to dry for 2 to 5 min. Antibiotic discs were fixed onto the agar plates by using sterilized forceps and were incubated at 37 °C for 24 h. The clear zone for each antibiotic disc was determined by measuring the diameter of the inhibition zone around the antibiotic disc.
4.3. Multiple Antibiotic Resistance (MAR) Index
The MAR index of the isolates against the tested antibiotics was calculated based on the following formula [17]. MAR index (multiple antibiotic resistance) = X/(Y × Z); where X = total number of antibiotic resistance cases; Y = total number of antibiotics used in the study; and Z = total number of bacterial isolates. A MAR index value of equal or less than 0.2 was defined as those antibiotics that were rarely or never used for the animal in terms of treatment; however, if the MAR index value was higher than 0.2, this was considered as an indicator of the high risk of exposure to those antibiotics received by the animals.
4.4. Plasmid Analysis
V. vulnificus strains were grown overnight in 10 mL of Lauria Bertani (LB) broth with an addition of 3% (w/v) of sodium chloride (NaCl) at 37 °C with shaking 200 rpm. A quantity of 1 mL of culture was then transferred into a 1.5 mL centrifuge tube and spun for 1 min at 10,000 rpm via a benchtop centrifuge (Minispin, Eppendorf, Germany). Plasmid extractions were conducted using PureYield™ Plasmid Miniprep System (Promega, Madison, WI, USA) as described in the manufacturer’s instructions. DNA plasmid was then analyzed on 1% (w/v) agarose gel. The gel was electrophoresed at 85 V for 1 h and Lambda DNA-HindIII Digested DNA was used as a DNA ladder (New England BioLabs, Ipswich, MA, USA). The gel was visualized using gel documentation (Syngene, Frederick, MD, USA).
4.5. RAPD Fingerprinting
A random amplified polymorphism DNA-polymerase chain reaction (RAPD-PCR) was used for characterization of the V. vulnificus isolated isolates. The RAPD11 primer used was 5′-AAAGCTGCGG-3′ and RAPD15 5′-CACACTCCAG-3′. The PCR technique was carried out in 0.2-μL microfuge tubes. The total volume of the reaction mixture was 50 μL, consisting of 25 μL 10× PCR master mix (EconoTaq®PULS GREEN 2X Master Mix, Lucigen, Middleton, WI, USA), 0.5 μL of OPC primer, and 1.0 μL (10–20 ng) of template DNA, the volume was then adjusted to a final volume by adding Nuclease Free Water (NFW). Concerning the negative control, one of the reaction mixtures without the DNA template was used. The solution mixture was placed in the Thermal Cycler (Bio-Rad, Hercules, CA, USA) and the PCR cycles parameters were denatured at 94 °C for 5 min followed by 45 cycles of denaturation at 94 °C for 1 min, annealing at 34 °C for 1 min, and polymerization at 72 °C for 2 min. Final elongation was carried out at 72 °C for 7 min. The 1 kb DNA ladder (Vivantis, Selangor, Malaysia) was used as a DNA size marker and fragments were viewed using a UV transilluminator (Syngene, Cambridge, UK).
4.6. Phylogenetic Analysis
Clonal relatedness of the V. vulnificus using primers RAPD11 and RAPD15 were analyzed as described by Sahilah et al. [26]. The image of the gel was analyzed using Gel ComparII (Applied Math, Kortjik, Belgium).
5. Conclusions
In conclusion, a high MAR index of V. vulnificus isolates in the study area indicated that the isolates originated from high-risk sources of contamination. No conclusion can be drawn as to the existence of R plasmids since no conjugation analysis was performed. However, in the present study, there was evidence that the resistant gene for certain antibiotics may be positioned either in plasmids or in genome DNA. While subtyping would increase the degree of strains differentiation and molecular approaches of RAPD, it is useful to differentiate V. vulnificus isolates, and thus increase the heterogeneity level of these isolates.
6. Patents
No patents have resulted from the work reported in this manuscript.
Acknowledgments
The authors wish to express their gratitude to grant ST-2014-016 and AP2017-002/1 for financial support. The gratitude is also extended to Tosiah Sadi and Zaimawati Md. Nejis from Malaysia Agriculture Research and Development Institute (MARDI) for their assistance in this research.
Supplementary Materials
The following are available online at https://www.mdpi.com/2079-6382/8/2/68/s1, Figure S1: Plasmid identification using PureYield™ Plasmid Miniprep System kit (Promega, USA) on 1% (w/v) agarose gel. Lane 1: Lambda. DNA-HindIII Digest DNA ladder. Lane 1-15: Vv isolates 1-15; Figure S2: Plasmid identification using PureYield™ Plasmid Miniprep System kit (Promega, USA) on 1%(w/v) agarose gel. Lane 1: Lambda DNA-HindIII Digest DNA ladder. Lane 2-11: Vv isolates 16-26. Figure S3: Plasmid identification using PureYield™ Plasmid Miniprep System kit (Promega, USA) on 1%(w/v) agarose gel. Lane 1: Lambda DNA-HindIII Digest DNA ladder. Lane 2- 6 Vv isolates 27-31. Figure S4: Plasmid identification using PureYield™ Plasmid Miniprep System kit (Promega, USA) on 1%(w/v) agarose gel. Lane 1: Lambda DNA-HindIII Digest DNA ladder. Lane 2-15: Vv isolates from 32 to 46 and lane 16: negative control. Figure S5: Plasmid identification using PureYield™ Plasmid Miniprep Systemkit (Promega, USA) on 1%(w/v) agarose gel. Lane M: Lambda DNA-HindIII Digest DNA ladder. Lane 2-15: Vv iolates from 47 to 60 and lane 16 NC: negative control. Table S1: Antibiotic resistance among Vibrio vulnificus isolates; Table S2: Antibiotic resistance among Vibrio vulnificus isolates.
Author Contributions
Conceptualization, S.A.M. and M.A.G.; Methodology, S.A.M.; Validation, N.A.M.Z.; Formal Analysis, S.A.M and M.M.K.A.-D.; Investigation, M.M.K.A.-D.; Resources, S.A.M.; Data Curation, M.M.K.A.-D.; Writing—Original Draft Preparation, M.M.K.A.-D.; Writing—Review and Editing A.A.A., N.A.M.Z., and S.A.M.; Supervision, S.A.M. and M.A.G.; Project Administration, S.A.M.; Funding Acquisition, S.A.M.
Funding
This research was funded by the grant of ST-2014-016 and AP2017-002/1.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Miyasaka J., Yahiro S., Arahira Y., Tokunaga H., Katsuk K., Hara-Kudo Y. Isolation of Vibrio parahaemolyticus and Vibrio vulnificus from wild aquatic birds in Japan. Epidemiol. Infect. 2006;134:780–785. doi: 10.1017/S0950268805005674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mohammed M.K.A.A., Sahilah A.M., Ma’aruf A.G., Mohammed A. A review of important virulence factors of Vibrio vulnificus. Curr. Res. J. Biol. Sci. 2014;6:76–88. [Google Scholar]
- 3.Paydar M., Thong K.L. Prevalence and genetic characterization of Vibrio vulnificus in raw seafood and seawater in Malaysia. J. Food Prot. 2013;76:1797–1800. doi: 10.4315/0362-028X.JFP-13-141. [DOI] [PubMed] [Google Scholar]
- 4.Sahilah A.M., Laila R.A., Sallehuddin H.M., Osman H., Aminah A., Ahmad Azuhairi A. Antibiotic resistance and molecular typing among cockle (Anadara granosa) strains of Vibrio parahaemolyticus by polymerase chain reaction (PCR)-based analysis. World J. Microbiol. Biotechnol. 2014;30:649–659. doi: 10.1007/s11274-013-1494-y. [DOI] [PubMed] [Google Scholar]
- 5.Antibiotic Resistance. [(accessed on 25 January 2019)]; Available online: https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance.
- 6.Srinivasan P., Ramasamy P. Occurrence, distribution and antibiotic resistance patterns of Vibrio species associated with viral diseased shrimp of south Indian Aquaculture environment. Int. J. Agric. Sci. 2009;1:1–10. [Google Scholar]
- 7.Lechumanan V., Yin W.F., Lee L.H., Chan K.G. Prevelence and antimicrobial susceptibility of Vibrio parahaemolyticus o isolated from retail shrimps in Malaysia. Front. Microbiol. 2015;6:33. doi: 10.3389/fmicb.2015.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lechumanan V., Pusparajah P., Tan L.T.H., Yin W.F., Lee L.H., Chan K.G. Occurence and antibiotic resistance of Vibrio parahaemolyticus from shellfish in Selangor, Malaysia. Front. Microbiol. 2015;6:1417. doi: 10.3389/fmicb.2015.01417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zulkifli Y., Alitheen N.B., Son R., Raha A.R., Samuel L., Yeap S.K., Nishibuchi M. Random amplified polymorphic DNA-PCR and ERIC PCR analysis on Vibrio parahaemolyticus isolated from cockles in Padang, Indonesia. Int. Food Res. J. 2009;16:141–150. [Google Scholar]
- 10.Roig F.J., Amaro C. Plasmid diversity in Vibrio vulnificus biotypes. Microbiology. 2009;155:489–497. doi: 10.1099/mic.0.023424-0. [DOI] [PubMed] [Google Scholar]
- 11.Elhadi N. Antibiotic Resistance and Plasmid Profiling of Clinically Significant Vibrio vulnificus Isolated from Coastal Water in Eastern Province of Saudi Arabia. Br. J. Pharm. Toxicol. 2012;3:93–97. [Google Scholar]
- 12.Radu S., Elhadi N.H.Z., Rusul G., Lihan S., Fifadara N., Yuherman, Purwati E. Characterization of Vibrio vulnificus isolated from cockles (Anadara granosa): Antimicrobial resistance, plasmid profiles and random amplification of polymorphic DNA analysis. FEMS Microbiol. Lett. 1998;165:139–143. doi: 10.1111/j.1574-6968.1998.tb13138.x. [DOI] [PubMed] [Google Scholar]
- 13.Radu S., Vincent M., Apun K., Rahim R.A., Benjamin P.G., Yuherman, Rusul G. Molecular characterization of Vibrio cholerae O1 outbreak strains in Miri, Sarawak (Malaysia) Acta Trop. 2002;83:168–176. doi: 10.1016/S0001-706X(02)00110-9. [DOI] [PubMed] [Google Scholar]
- 14.Okoh A.I., Igbinosa E.O. Antibiotic susceptibility profiles of some Vibrio strains isolated from wastewater final effluents in a rural community of Eastern Cape Province of South Africa. BMC Microbiol. 2010;10:143. doi: 10.1186/1471-2180-10-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baker-Austin C., Stockley L., Rangdale R., Martinez-Urtaza J. Environmental occurrence and clinical impact of Vibrio vulnificus and Vibrio parahaemolyticus: A European perspective. Environ. Microbiol. Rep. 2010;2:7–18. doi: 10.1111/j.1758-2229.2009.00096.x. [DOI] [PubMed] [Google Scholar]
- 16.Tunung R., Jeyaletchumi P., Noorlis A., Tang Y.H., Sandra A., Ghazali F.M., Noranizan M.A., Lesley M.B., Haresh K.K., Nakaguchi Y., et al. Biosafety of Vibrio parahaemolyticus from vegetables based on antimicrobial sensitivity and RAPD profiling. Int. Food Res. J. 2012;19:467–474. [Google Scholar]
- 17.Lee S.W., Wendy W. Antibiogram and Heavy Metal Resistance Pattern of Salmonella spp. Isolated from Wild Asian Sea Bass (Lates calcarifer) from Tok Bali, Kelantan, Malaysia. Jordan J. Biol. Sci. 2011;4:125–128. [Google Scholar]
- 18.Dale J.W., Park S. Molecular Genetics of Bacteria. 1st ed. John Wiley & Sons Inc.; Chichester, UK: 2010. pp. 147–148. [Google Scholar]
- 19.Guglielgmetti E., Korhonen J.M., Haikkinen J., Morelli L., Von Wright A. Transfer and plasmid-mediated resistance to tetracycline in pathogenic bacteria from fish and aquaculture environments. FEMS Microbiol. Lett. 2009;293:28–34. doi: 10.1111/j.1574-6968.2009.01512.x. [DOI] [PubMed] [Google Scholar]
- 20.Aoki T., Kitao T., Watanabe S., Takeshita S. Drug resistance and R plasmids in Vibrio anguillarum isolated in cultured ayu (Plecoglossus altivelis) Microbiol. Immunol. 1984;28:1–9. doi: 10.1111/j.1348-0421.1984.tb02942.x. [DOI] [PubMed] [Google Scholar]
- 21.Zhang R., Wang Y., Gu J.D. Identification of environmental plasmid-bearing Vibrio species isolated from polluted and pristine marine reserves of Hong Kong, and resistance to antibiotics and mercury. Antonic van Leeuwenhoek. 2006;89:307–315. doi: 10.1007/s10482-005-9032-z. [DOI] [PubMed] [Google Scholar]
- 22.Zhang R., Pan L., Zhao Z. High incidence of plasmids in marine Vibrio species isolated from Mai Po Nature Reserve of Hong Kong. Ecotoxicology. 2012;21:1661–1668. doi: 10.1007/s10646-012-0939-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sahilah A.M., Audrey L.Y.Y., Ong S.L., Wan Sakeenah W.N., Safiyyah S., Norrakiah A.S., Aminah A., Ahmad Azuhairi A. DNA profiling among egg and beaf meat isolates of Escherichia coli by enterobacterial repetitive intergenic consensus-PCR (ERIC-PCR) and random amplified polymorphic DNA-PCR (RAPD-PCR) Int. Food Res. J. 2010;17:853–866. [Google Scholar]
- 24.Nisreen J.K., Sahilah A.M. Genetic diversity of Bacillus cereus isolated from fried rice. Int. J. ChemTech Res. 2018;11:52–59. [Google Scholar]
- 25.Bauer A.W., Kirby W.M.M., Sherris J.C., Turl M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966;45:493–496. doi: 10.1093/ajcp/45.4_ts.493. [DOI] [PubMed] [Google Scholar]
- 26.Sahilah A.M., Son R., Rusul G., Samuel L., Hassan Z., Lum K.Y., Ahmad M.A. Molecular typing of Salmonella weltevreden and Salmonella chincol by pulsed field gel electrophoresis (PFGE) and enterobacterial repetitive intergenic consensuspolymerase chain reaction (ERIC-PCR) World J. Microbiol. Biotechnol. 2000;16:621–624. doi: 10.1023/A:1008967914499. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.