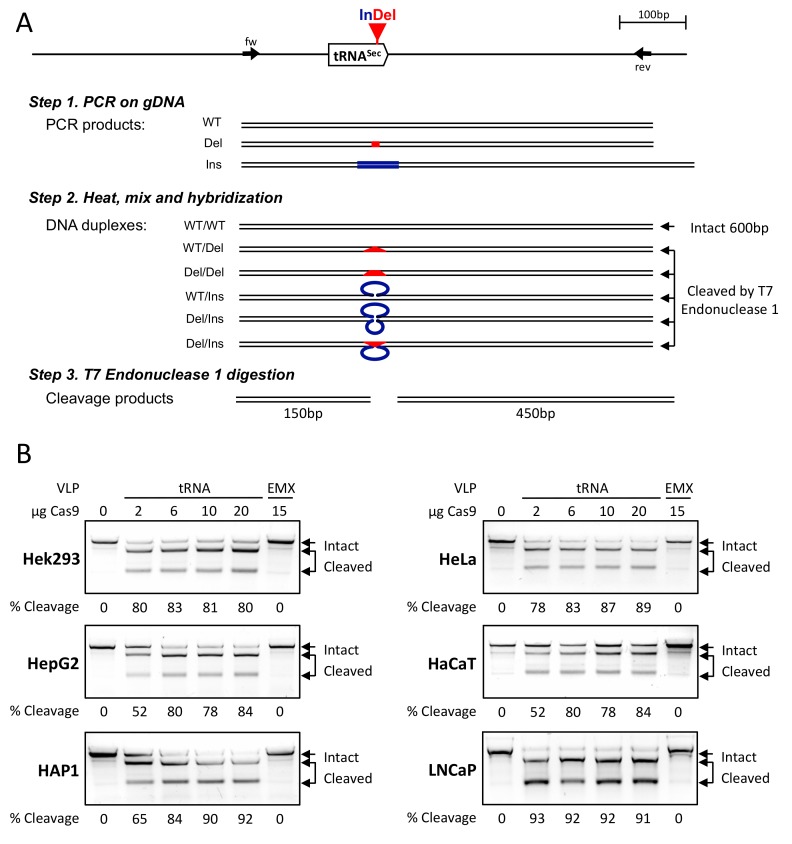
Figure 3.
Evaluation of tRNA[Ser]Sec gene editing activities by CRISPR-Cas9-VLPs used in various cell lines with T7 endonuclease 1 mismatch assay. (A) Illustration of this assay applied to detect Cas9 induced mutations. The gDNA was extracted from cells treated with Cas9-sgRNA and used for a polymerase chain reaction (PCR) amplification of the tRNA[Ser]Sec gene using forward (fw) and reverse (rev) primers. The PCR product (fragment of approximately 600 bp) is composed of a mixture between the original wild-type (WT) and the Cas9 induced mutated sequences. The deletions (Del) and insertions (Ins) are represented in red and blue, respectively. The PCR products were heated, mixed and re-hybridized before addition of the T7 endonuclease 1, which cuts when heteroduplexes are formed during annealing (i.e., WT/Del, Del/Del, WT/Ins, Del/Ins and Del/Ins). The size of the digestion products were 150 and 450 bp. (B) The six different cell lines were treated with increasing amount CRISPR-Cas9-VLPs, targeting either tRNA[Ser]Sec or EMX gene. Four days post-treatment, the cells were harvested, and their gDNA was analyzed with T7 endonuclease 1 assay. An aliquot of the reaction was run on an 2.5% agarose gel. The cleavage efficiency of the PCR fragment was calculated as the percentage of cleaved products relative to all fragments in each lane. The positions of the intact and cleaved DNAs are indicated by arrows.